
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,670,138 – 4,670,378 |
| Length | 240 |
| Max. P | 0.774608 |

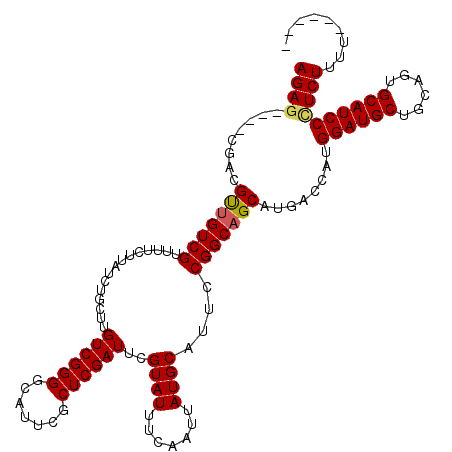
| Location | 4,670,138 – 4,670,258 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.05 |
| Mean single sequence MFE | -33.08 |
| Consensus MFE | -27.16 |
| Energy contribution | -27.35 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.82 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4670138 120 + 22224390 AGAGAGAGAGACGCUGUCGUUUUCUUAUCUGCUUGUCGGGGCAUUCGCUCGAUUCGUAUUUCAAUUAUGCAUUCCGGCAGCAUGACCAUGGAUGCUGCAGUGCAUCCCUCUUUCCCCACU ...(((((((..(((((((...............((((((.......))))))..((((.......))))....)))))))........((((((......)))))))))))))...... ( -37.30) >DroSec_CAF1 30895 111 + 1 AGAG----CGACGUUGUCGUUUUCUUAUCUGCUUGUCGGGGCAUUCGCUCGAUUCGUAUUUCAAUUAUGCAUUCCGGCAGCAUGACCAUGGAUGCUGCAGUGCAUCCCUCUUUUU----- ((((----((((...))))))))....((((((.((((((((....)))).....((((.......))))....)))))))).))....((((((......))))))........----- ( -32.00) >DroEre_CAF1 31591 110 + 1 AGAG----CGACGUUGUCGUUUUCUUAUCUUCUUGUCGGGGCAUUCGCUCGAUUCGUAUUUCAAUUAUGCAUUCCGGCAGCACGACCAUGGAUGCUGCAGUGCAUCCGUCUUUU------ ....----.((((((((((...............((((((.......))))))..((((.......))))....)))))))........((((((......)))))))))....------ ( -30.60) >DroYak_CAF1 28419 110 + 1 AGAG----CGACGUCGUCGUUUUCUUAUCUGCUUGUCGGGGCAUUCGCUCGAUUCGUAUUUCAAUUAUGCAUUCCGGCAGCAUGACCAUGGAUGCUGCAGUGCAUCCUUCUUUU------ ((((----((((...))))))))....((((((.((((((((....)))).....((((.......))))....)))))))).))....((((((......)))))).......------ ( -32.40) >consensus AGAG____CGACGUUGUCGUUUUCUUAUCUGCUUGUCGGGGCAUUCGCUCGAUUCGUAUUUCAAUUAUGCAUUCCGGCAGCAUGACCAUGGAUGCUGCAGUGCAUCCCUCUUUU______ ((((........(((((((...............((((((.......))))))..((((.......))))....)))))))........((((((......))))))))))......... (-27.16 = -27.35 + 0.19)


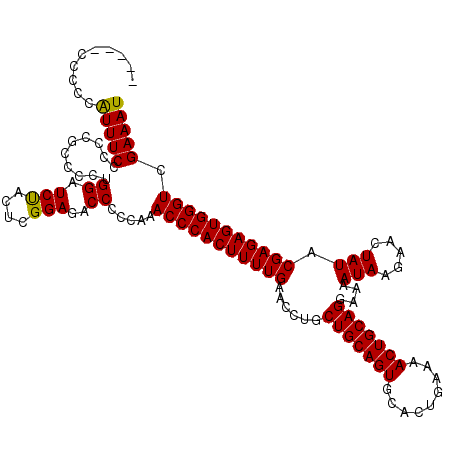
| Location | 4,670,258 – 4,670,378 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.21 |
| Mean single sequence MFE | -31.42 |
| Consensus MFE | -29.25 |
| Energy contribution | -28.88 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.774608 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4670258 120 + 22224390 CCCCCCCCCCAUUUCCCCCGUCCCUGGAUCUACACGGAGACCCCCAAACCCACUUUUGAACCUGCUGCAGUGCACUGAAAACUGCAGGAAAAUAAGAACUAUACGAGAGUGGGUCGAAAU ..........(((((....(((((((......)).)).)))......(((((((((((......(((((((.........)))))))....(((.....))).))))))))))).))))) ( -32.50) >DroSec_CAF1 31006 116 + 1 ----UCCCCCAUUUCCCCCGCCCCUGGAUCUACUCGGAGACCCCCAAACCCACUUUUGAACCUGCUGCAGUGCACUGGAAACUGCAGGAAAAUAAGAACUAUACGAGAGUGGGUCGAAAU ----......(((((..........((.(((......))).))....(((((((((((......(((((((.........)))))))....(((.....))).))))))))))).))))) ( -29.30) >DroEre_CAF1 31701 115 + 1 -----GCUCCGUUUCCCCUGCCCCUGGAUCUACUCGGAGACCCCCAAACCCACUUUUGAACCUGCUGCAGUGCACUGAAAACUGCAGGAAAAUAAGAGCUAUACGAGAGUGGGUCGAAAU -----.(((((..(((.........)))......)))))........(((((((((((......(((((((.........)))))))....(((.....))).)))))))))))...... ( -33.00) >DroYak_CAF1 28529 115 + 1 -----CCUCCAUUUCUCCCGCCCCUGGAUCCACUCGGAGACCCCCAAACCCACUUUUGAACCUGCUGCAGUGCACUGAAAACUGCAGGAAAAUAAGAACUAUACGAGAGUGGGUCGAAAU -----.....(((((..........((.(((....)))..)).....(((((((((((......(((((((.........)))))))....(((.....))).))))))))))).))))) ( -30.90) >consensus _____CCCCCAUUUCCCCCGCCCCUGGAUCUACUCGGAGACCCCCAAACCCACUUUUGAACCUGCUGCAGUGCACUGAAAACUGCAGGAAAAUAAGAACUAUACGAGAGUGGGUCGAAAU ..........(((((..........((.(((....)))..)).....(((((((((((......(((((((.........)))))))....(((.....))).))))))))))).))))) (-29.25 = -28.88 + -0.37)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:15:56 2006