
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,667,136 – 4,667,394 |
| Length | 258 |
| Max. P | 0.932320 |

| Location | 4,667,136 – 4,667,240 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 97.44 |
| Mean single sequence MFE | -30.47 |
| Consensus MFE | -27.93 |
| Energy contribution | -28.60 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.655647 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4667136 104 + 22224390 GUGGCAUCGCGGCUUUGUGUGCUGCGCUGUUUGUUUAGCGUUCCUUAAGCUCUUAAGCUCAUUUCGGUUGCGUUUUAUUUGUAACUUGAGGCACCCUACACACA ((((((.((((((.......)))))).))).....(((.((.(((((((((....))))......(((((((.......)))))))))))).)).))))))... ( -30.60) >DroSec_CAF1 28413 102 + 1 GUGGCAUCGCGGCUUUGUGUGCUGCGCUGUUUGUUUAGCGUUGCUUAAGCUCUUAAGCUCAUUUCGGUUGCGUUUUAUUUGUAACUUGAAGCACCCUACACA-- ((((((.((((((.......)))))).))).......((...((((((....))))))....((((((((((.......)))))).))))))......))).-- ( -30.20) >DroEre_CAF1 29158 104 + 1 GUGGCAUCGCGGCUUUGUGUGCUGCGCUGUUUGUUUAGCGUUCCUUAAGCUCUUAAGCUCAUUUCGGUUGCGUUUUAUUUGUAACUUGAGGCACCCUACACACA ((((((.((((((.......)))))).))).....(((.((.(((((((((....))))......(((((((.......)))))))))))).)).))))))... ( -30.60) >consensus GUGGCAUCGCGGCUUUGUGUGCUGCGCUGUUUGUUUAGCGUUCCUUAAGCUCUUAAGCUCAUUUCGGUUGCGUUUUAUUUGUAACUUGAGGCACCCUACACACA ((((((.((((((.......)))))).))).....(((.((.(((((((((....))))......(((((((.......)))))))))))).)).))))))... (-27.93 = -28.60 + 0.67)

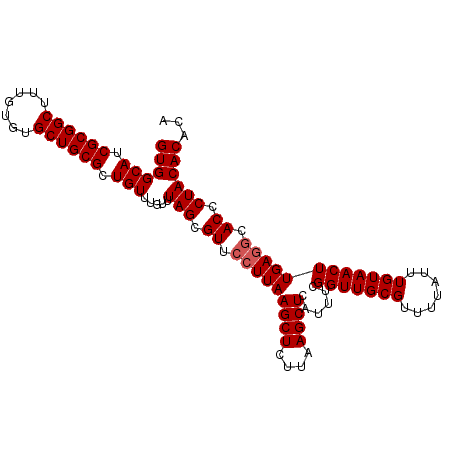

| Location | 4,667,176 – 4,667,280 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 92.95 |
| Mean single sequence MFE | -28.60 |
| Consensus MFE | -23.35 |
| Energy contribution | -24.69 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.774763 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4667176 104 - 22224390 AUAAAGUCAGCUUUGGUGUGAGAAUUUCGGUGUGUGUGUGUGUGUGUAGGGUGCCUCAAGUUACAAAUAAAACGCAACCGAAAUGAGCUUAAGAGCUUAAGGAA ........((((((...((....((((((((.(((((...(((.(((((..((...))..))))).)))..)))))))))))))..))...))))))....... ( -28.30) >DroSec_CAF1 28453 96 - 1 AUAAAGUCAGCUUUGGUGUGAGUAUUUCGGUGUG--------UGUGUAGGGUGCUUCAAGUUACAAAUAAAACGCAACCGAAAUGAGCUUAAGAGCUUAAGCAA .........((((.(((.((((((((((((((((--------(.(((((..((...))..)))))......)))).))))))))..)))))...))).)))).. ( -27.90) >DroEre_CAF1 29198 104 - 1 AUAAAGUCAGCUUUGGUGUGAGUAUUUCGGUGUGUGUGUGUGUGUGUAGGGUGCCUCAAGUUACAAAUAAAACGCAACCGAAAUGAGCUUAAGAGCUUAAGGAA ........((((((...((...(((((((((.(((((...(((.(((((..((...))..))))).)))..)))))))))))))).))...))))))....... ( -29.60) >consensus AUAAAGUCAGCUUUGGUGUGAGUAUUUCGGUGUGUGUGUGUGUGUGUAGGGUGCCUCAAGUUACAAAUAAAACGCAACCGAAAUGAGCUUAAGAGCUUAAGGAA ........((((((...((...(((((((((.(((((.......(((((..((...))..)))))......)))))))))))))).))...))))))....... (-23.35 = -24.69 + 1.33)



| Location | 4,667,240 – 4,667,360 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.58 |
| Mean single sequence MFE | -19.38 |
| Consensus MFE | -18.70 |
| Energy contribution | -19.07 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932320 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4667240 120 + 22224390 CACACACACACCGAAAUUCUCACACCAAAGCUGACUUUAUUUAUUUUAAAUAAAGCUGCCAAGCAGUUGAAAUAUUGAGUCAAACAAUUGCAAGACGACAGCUGCAGCUUCAAAAUUAAA ...........................(((((..(((((((((...))))))))).......(((((((.....((((((......))).))).....)))))))))))).......... ( -19.40) >DroSec_CAF1 28515 114 + 1 ------CACACCGAAAUACUCACACCAAAGCUGACUUUAUUUAUUUUAAAUAAAGCUGCCAAGCAGAUGAAAUAUUGAGUCAAACAAUUGCAAGACGACAGCUGCAGCUUCAAAAUUAAA ------.....................((((((((((....(((((((.......((((...)))).)))))))..))))))......((((.(.(....)))))))))).......... ( -16.70) >DroEre_CAF1 29262 120 + 1 CACACACACACCGAAAUACUCACACCAAAGCUGACUUUAUUUAUUUUAAAUAAAGCUGCAAAGCAGCUAAAAUAUUGAGUCCAACAAUUGCAAGACGACAGCUGCAGCUUCAAAAUUAAA ............(((..............((.(.(((((((((...)))))))))).))...((((((......((((((......))).)))......))))))...)))......... ( -19.20) >DroYak_CAF1 25760 120 + 1 CACACACCCACCGAAAUACUCACACCAAAGCUGACUUUAUUUAUUUUAAAUAAAGCUGCAAAGCAGCUGAAAUAUUGAGUCAAACAAUUGCAAGACGACAGCUGCAGCUUCAAAAUUAAA ............(((..............((.(.(((((((((...)))))))))).))...(((((((.....((((((......))).))).....)))))))...)))......... ( -22.20) >consensus CACACACACACCGAAAUACUCACACCAAAGCUGACUUUAUUUAUUUUAAAUAAAGCUGCAAAGCAGCUGAAAUAUUGAGUCAAACAAUUGCAAGACGACAGCUGCAGCUUCAAAAUUAAA ...........................(((((..(((((((((...))))))))).......(((((((.....((((((......))).))).....)))))))))))).......... (-18.70 = -19.07 + 0.37)


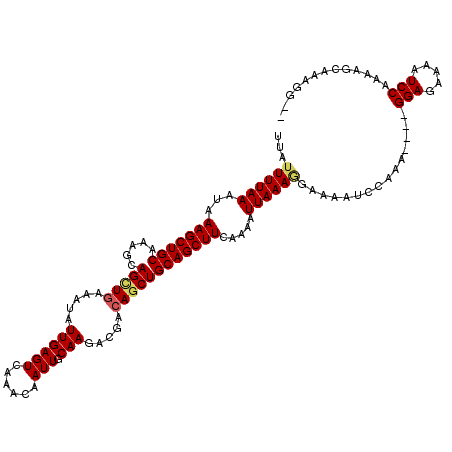
| Location | 4,667,280 – 4,667,394 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.64 |
| Mean single sequence MFE | -19.99 |
| Consensus MFE | -15.07 |
| Energy contribution | -15.57 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.708095 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4667280 114 + 22224390 UUAUUUUAAAUAAAGCUGCCAAGCAGUUGAAAUAUUGAGUCAAACAAUUGCAAGACGACAGCUGCAGCUUCAAAAUUAAAGUAAAAUCCAAA----GGAGAAAAUCCAAGAGCAAAGG-- (((((((((...(((((((...(((((((...............))))))).((.(....)))))))))).....)))))))))........----(((.....)))...........-- ( -22.46) >DroSec_CAF1 28549 118 + 1 UUAUUUUAAAUAAAGCUGCCAAGCAGAUGAAAUAUUGAGUCAAACAAUUGCAAGACGACAGCUGCAGCUUCAAAAUUAAAAGAAAAUCCAAAAGGAGGAGAAAAUCCAAGAGCAAAGG-- ...((((((...(((((((..(((.((((...))))..(((............)))....)))))))))).....)))))).....(((....)))(((.....)))...........-- ( -18.60) >DroEre_CAF1 29302 116 + 1 UUAUUUUAAAUAAAGCUGCAAAGCAGCUAAAAUAUUGAGUCCAACAAUUGCAAGACGACAGCUGCAGCUUCAAAAUUAAACCAAAAUCCAAA----GGAGAAAAUCCAAAACCAAAGUAU .((((((.....((((((((.....(((......((((((......))).)))......)))))))))))......................----(((.....)))......)))))). ( -16.80) >DroYak_CAF1 25800 116 + 1 UUAUUUUAAAUAAAGCUGCAAAGCAGCUGAAAUAUUGAGUCAAACAAUUGCAAGACGACAGCUGCAGCUUCAAAAUUAAAGGAAAAUCCAAA----GGACAAAAUCCAAAAGCAAAGUAU .((((((...............(((((((.....((((((......))).))).....))))))).((((..........((.....))...----(((.....)))..)))))))))). ( -22.10) >consensus UUAUUUUAAAUAAAGCUGCAAAGCAGCUGAAAUAUUGAGUCAAACAAUUGCAAGACGACAGCUGCAGCUUCAAAAUUAAAGGAAAAUCCAAA____GGAGAAAAUCCAAAAGCAAAGG__ ...((((((...(((((((.....(((((.....((((((......))).))).....)))))))))))).....))))))...............(((.....)))............. (-15.07 = -15.57 + 0.50)

| Location | 4,667,280 – 4,667,394 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.64 |
| Mean single sequence MFE | -24.05 |
| Consensus MFE | -18.96 |
| Energy contribution | -18.96 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.863697 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4667280 114 - 22224390 --CCUUUGCUCUUGGAUUUUCUCC----UUUGGAUUUUACUUUAAUUUUGAAGCUGCAGCUGUCGUCUUGCAAUUGUUUGACUCAAUAUUUCAACUGCUUGGCAGCUUUAUUUAAAAUAA --.......(((.(((.....)))----...)))......(((((...((((((((((((.((.(((..((....))..)))...........)).)))..))))))))).))))).... ( -21.70) >DroSec_CAF1 28549 118 - 1 --CCUUUGCUCUUGGAUUUUCUCCUCCUUUUGGAUUUUCUUUUAAUUUUGAAGCUGCAGCUGUCGUCUUGCAAUUGUUUGACUCAAUAUUUCAUCUGCUUGGCAGCUUUAUUUAAAAUAA --...........(((.....)))(((....))).....((((((...(((((((((.......(((..((....))..))).........((......))))))))))).))))))... ( -23.40) >DroEre_CAF1 29302 116 - 1 AUACUUUGGUUUUGGAUUUUCUCC----UUUGGAUUUUGGUUUAAUUUUGAAGCUGCAGCUGUCGUCUUGCAAUUGUUGGACUCAAUAUUUUAGCUGCUUUGCAGCUUUAUUUAAAAUAA ..(((..(((((.(((.....)))----...)))))..)))((((...((((((((((((((..((((.((....)).)))).........)))).....)))))))))).))))..... ( -24.50) >DroYak_CAF1 25800 116 - 1 AUACUUUGCUUUUGGAUUUUGUCC----UUUGGAUUUUCCUUUAAUUUUGAAGCUGCAGCUGUCGUCUUGCAAUUGUUUGACUCAAUAUUUCAGCUGCUUUGCAGCUUUAUUUAAAAUAA .............(((....((((----...))))..)))(((((...((((((((((((((..(((..((....))..))).........)))).....)))))))))).))))).... ( -26.60) >consensus __ACUUUGCUCUUGGAUUUUCUCC____UUUGGAUUUUCCUUUAAUUUUGAAGCUGCAGCUGUCGUCUUGCAAUUGUUUGACUCAAUAUUUCAGCUGCUUGGCAGCUUUAUUUAAAAUAA ...((........(((.....))).......)).......(((((...((((((((((((((..(((..((....))..))).))...........)))..))))))))).))))).... (-18.96 = -18.96 + 0.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:15:51 2006