
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,666,070 – 4,666,197 |
| Length | 127 |
| Max. P | 0.876934 |

| Location | 4,666,070 – 4,666,166 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.69 |
| Mean single sequence MFE | -23.87 |
| Consensus MFE | -14.23 |
| Energy contribution | -14.98 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.876934 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4666070 96 + 22224390 GCUAAUCGAUGAGC---------------------AAAUAGUGCACUCGUCUAAUCGAUGACUUUCUGAUAGCCUCGAUUGCACUUC---CUUCACUUAUUUCCCAGCCCAAUCGCUGCU .......((((((.---------------------....((((((.(((.(((.((((......)).)))))...))).))))))..---.....))))))...((((......)))).. ( -19.60) >DroSec_CAF1 27398 90 + 1 GCUAAUCGAUUGGC---------------------AAAUAGUGCACUCGUCUAAUCGAUGACUUUCUGAUAGUCUCGAUUCCCCCUC---C------UAUUUCCCAGUCCAAUCGCUGCU ((.....((((((.---------------------((((((.(.........((((((.((((.......))))))))))......)---)------))))).)))))).....)).... ( -20.56) >DroEre_CAF1 28135 99 + 1 GCUAAUCGAUUGGC---------------------AAAUAGUGCACUCGUCUAAUCGAUGACUUUCUGAUAGUCUCGAUUGCACUUCUACUCCUACUUAGUGCCCACCCCAAUUGCUGCU ((....(((((((.---------------------.................((((((.((((.......))))))))))(((((.............))))).....)))))))..)). ( -22.62) >DroYak_CAF1 24605 118 + 1 GCUAAUCGAUUGGUAAAUGGCAAAUGGUAAAUGGUAAAUAGUGCACUAUUCUAAUCGAUGACUUUCUGAUAGUCUCGAUUGCACUU--GCCUCUACUUAUUUCCCGCCCCAAUUGCUGCU ((....(((((((....(((.((((((((...((((...((((((........(((((.((((.......))))))))))))))))--)))..))).))))).)))..)))))))..)). ( -32.70) >consensus GCUAAUCGAUUGGC_____________________AAAUAGUGCACUCGUCUAAUCGAUGACUUUCUGAUAGUCUCGAUUGCACUUC___CUCUACUUAUUUCCCACCCCAAUCGCUGCU ((....(((((((..........................((....))....(((((((.((((.......)))))))))))...........................)))))))..)). (-14.23 = -14.98 + 0.75)


| Location | 4,666,089 – 4,666,197 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 87.44 |
| Mean single sequence MFE | -26.70 |
| Consensus MFE | -19.15 |
| Energy contribution | -19.65 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.753638 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
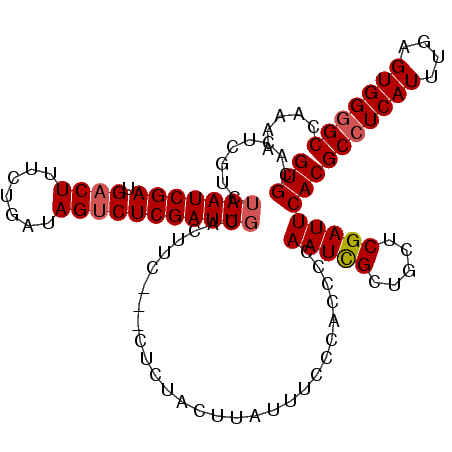
>X_DroMel_CAF1 4666089 108 + 22224390 GUGCACUCGUCUAAUCGAUGACUUUCUGAUAGCCUCGAUUGCACUUC---CUUCACUUAUUUCCCAGCCCAAUCGCUGCUCGAUUCACGCCUCAUUUGAGUGGGGCCAAAA (((((.(((.(((.((((......)).)))))...))).)))))...---..............((((......))))..........(((((((....)))))))..... ( -25.60) >DroSec_CAF1 27417 102 + 1 GUGCACUCGUCUAAUCGAUGACUUUCUGAUAGUCUCGAUUCCCCCUC---C------UAUUUCCCAGUCCAAUCGCUGCUCGAUUCACGCCUCAUUUGAGUGGGGCCAAAA (((...(((...((((((.((((.......)))))))))).......---.------.......((((......))))..)))..)))(((((((....)))))))..... ( -24.80) >DroEre_CAF1 28154 110 + 1 GUGCACUCGUCUAAUCGAUGACUUUCUGAUAGUCUCGAUUGCACUUCUACUCCUACUUAGUGCCCACCCCAAUUGCUGCUCGAUUCACGCCUCAUUUGAGUGG-GCCAAAA (((((........(((((.((((.......))))))))))))))...............(.((((((..(((.((..((.........))..)).))).))))-))).... ( -28.60) >DroYak_CAF1 24645 109 + 1 GUGCACUAUUCUAAUCGAUGACUUUCUGAUAGUCUCGAUUGCACUU--GCCUCUACUUAUUUCCCGCCCCAAUUGCUGCUCGAUUCACGCCUCAUUUGAGUGGGGCCAAAA (((((........(((((.((((.......))))))))))))))..--(((.(((((((......((...(((((.....)))))...))......))))))))))..... ( -27.80) >consensus GUGCACUCGUCUAAUCGAUGACUUUCUGAUAGUCUCGAUUGCACUUC___CUCUACUUAUUUCCCACCCCAAUCGCUGCUCGAUUCACGCCUCAUUUGAGUGGGGCCAAAA (((........(((((((.((((.......))))))))))).............................(((((.....))))))))(((((((....)))))))..... (-19.15 = -19.65 + 0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:15:47 2006