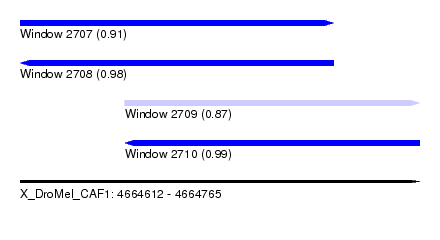
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,664,612 – 4,664,765 |
| Length | 153 |
| Max. P | 0.988247 |

| Location | 4,664,612 – 4,664,732 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.83 |
| Mean single sequence MFE | -26.45 |
| Consensus MFE | -23.64 |
| Energy contribution | -23.20 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.912548 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
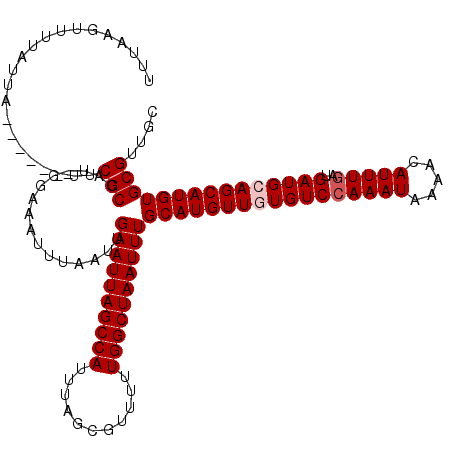
>X_DroMel_CAF1 4664612 120 + 22224390 AAACUAGGCUAAGCAAAAAGUGCUCGUUUAUUUGCACUCCGCAACGCACAUGCUGCAUCAUCAAAUGUUUUAUUUGGACACAACAUGCAAAUUAGCCAAAAACGCUAAAUGGCUAAUUCA ...(((((..(((((...(((((..........)))))..(((..((....))))).........)))))..)))))............(((((((((...........))))))))).. ( -23.90) >DroSec_CAF1 25945 120 + 1 AAACUGGGCUAAGCAAAAAGUGCUCGUUUAUUUGCACUCCGCAACGCACAUGCUGCAUCAUCAAAUGUUUUAUUUGGACACAACAUGCAAAUUAGCCAAAAACGCUAAAUGGCUAAUUCA ...(..((..(((((...(((((..........)))))..(((..((....))))).........)))))..))..)............(((((((((...........))))))))).. ( -23.70) >DroEre_CAF1 26700 120 + 1 AAACCAGGCUAUGCAAAAAGUGCACGUUUAUUUGCACUCCGCAACGCACAUGCUGCAUCAUCAAAUGUUUUAUUUGGACAAAACAUGCAAAUUAGCCAAAAACGCUAAAUGGCUAAUUCA .......((.(((((...((((((........))))))..(((.......))))))))......(((((((........))))))))).(((((((((...........))))))))).. ( -25.80) >DroYak_CAF1 23110 120 + 1 AAACUAGGCUGUGCAAAAAGUGCACGUUUAUUUGCAUUCCGCAGCGCACAUGCUGCAUCAUCAAAUGUUUUAUUUCGACAAAACAUGCAAAUUAGCCAAAAACGCUAAAUGGCUAAUUCA ....(((((.(((((.....))))))))))(((((.....((((((....))))))........(((((((........))))))))))))(((((((...........))))))).... ( -32.40) >consensus AAACUAGGCUAAGCAAAAAGUGCACGUUUAUUUGCACUCCGCAACGCACAUGCUGCAUCAUCAAAUGUUUUAUUUGGACAAAACAUGCAAAUUAGCCAAAAACGCUAAAUGGCUAAUUCA ......(((((((((.....)))).....(((((((....(((..((....))))).........(((((.....))))).....))))))))))))......(((....)))....... (-23.64 = -23.20 + -0.44)

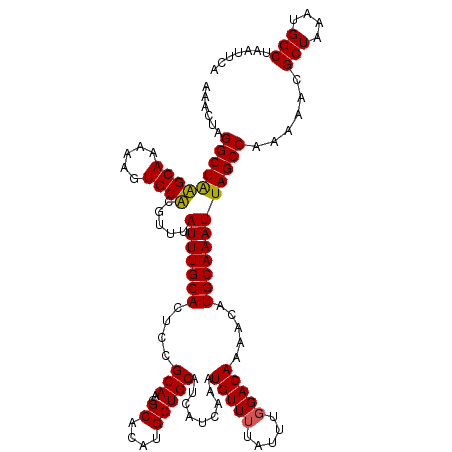

| Location | 4,664,612 – 4,664,732 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.83 |
| Mean single sequence MFE | -36.93 |
| Consensus MFE | -33.47 |
| Energy contribution | -34.48 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.14 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.977901 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4664612 120 - 22224390 UGAAUUAGCCAUUUAGCGUUUUUGGCUAAUUUGCAUGUUGUGUCCAAAUAAAACAUUUGAUGAUGCAGCAUGUGCGUUGCGGAGUGCAAAUAAACGAGCACUUUUUGCUUAGCCUAGUUU .((((((((((...........))))))))))(((((((((((((((((.....)))))..))))))))))))((...(((((((((..........))))))..)))...))....... ( -38.70) >DroSec_CAF1 25945 120 - 1 UGAAUUAGCCAUUUAGCGUUUUUGGCUAAUUUGCAUGUUGUGUCCAAAUAAAACAUUUGAUGAUGCAGCAUGUGCGUUGCGGAGUGCAAAUAAACGAGCACUUUUUGCUUAGCCCAGUUU .((((((((((...........))))))))))(((((((((((((((((.....)))))..))))))))))))((...(((((((((..........))))))..)))...))....... ( -38.70) >DroEre_CAF1 26700 120 - 1 UGAAUUAGCCAUUUAGCGUUUUUGGCUAAUUUGCAUGUUUUGUCCAAAUAAAACAUUUGAUGAUGCAGCAUGUGCGUUGCGGAGUGCAAAUAAACGUGCACUUUUUGCAUAGCCUGGUUU .((((((((((...........))))))))))(((((((.(((((((((.....)))))..)))).)))))))((..(((((((((((........)))))))..))))..))....... ( -34.70) >DroYak_CAF1 23110 120 - 1 UGAAUUAGCCAUUUAGCGUUUUUGGCUAAUUUGCAUGUUUUGUCGAAAUAAAACAUUUGAUGAUGCAGCAUGUGCGCUGCGGAAUGCAAAUAAACGUGCACUUUUUGCACAGCCUAGUUU .((((((((((...........))))))))))((..(((((((....)))))))(((((((..((((((......))))))..)).)))))....(((((.....))))).))....... ( -35.60) >consensus UGAAUUAGCCAUUUAGCGUUUUUGGCUAAUUUGCAUGUUGUGUCCAAAUAAAACAUUUGAUGAUGCAGCAUGUGCGUUGCGGAGUGCAAAUAAACGAGCACUUUUUGCAUAGCCUAGUUU .((((((((((...........))))))))))(((((((((((((((((.....)))))..))))))))))))((...(((((((((..........))))))..)))...))....... (-33.47 = -34.48 + 1.00)

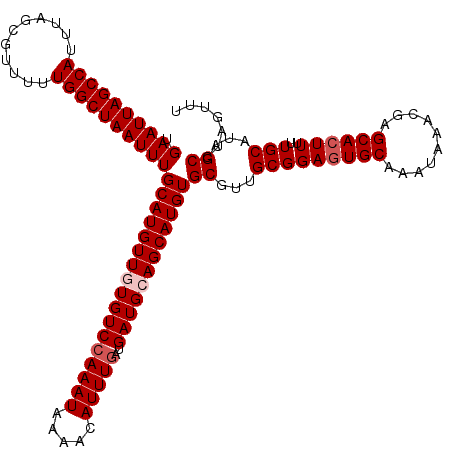
| Location | 4,664,652 – 4,664,765 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.56 |
| Mean single sequence MFE | -22.24 |
| Consensus MFE | -17.45 |
| Energy contribution | -17.70 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.874135 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4664652 113 + 22224390 GCAACGCACAUGCUGCAUCAUCAAAUGUUUUAUUUGGACACAACAUGCAAAUUAGCCAAAAACGCUAAAUGGCUAAUUCAAUUAAAUUUCC-AUGGCGAA------UAAUAGAACUUAAA ....(((.((((.(((((.......(((((.....)))))....)))))(((((((((...........)))))))))............)-))))))..------.............. ( -21.90) >DroSec_CAF1 25985 114 + 1 GCAACGCACAUGCUGCAUCAUCAAAUGUUUUAUUUGGACACAACAUGCAAAUUAGCCAAAAACGCUAAAUGGCUAAUUCAAUUAAAUUUCCAAUGGCGAA------UAAUAAAACUUAAA ....(((.(((..(((((.......(((((.....)))))....)))))(((((((((...........)))))))))..............))))))..------.............. ( -19.60) >DroEre_CAF1 26740 113 + 1 GCAACGCACAUGCUGCAUCAUCAAAUGUUUUAUUUGGACAAAACAUGCAAAUUAGCCAAAAACGCUAAAUGGCUAAUUCAAUUAAAUUUCU-AUGGCGAA------UAAUAAAACUUAGA ....(((.((((.(((((.......(((((.....)))))....)))))(((((((((...........)))))))))............)-))))))..------.............. ( -20.20) >DroYak_CAF1 23150 119 + 1 GCAGCGCACAUGCUGCAUCAUCAAAUGUUUUAUUUCGACAAAACAUGCAAAUUAGCCAAAAACGCUAAAUGGCUAAUUCAAUUAAAUUUCC-AUUGCGAGUUUCCACAAUGAAACUUAAA ((((((....))))))........(((((((........)))))))((((((((((((...........))))))))..............-.))))(((((((......)))))))... ( -27.26) >consensus GCAACGCACAUGCUGCAUCAUCAAAUGUUUUAUUUGGACAAAACAUGCAAAUUAGCCAAAAACGCUAAAUGGCUAAUUCAAUUAAAUUUCC_AUGGCGAA______UAAUAAAACUUAAA ....(((......(((((.......(((((.....)))))....)))))(((((((((...........))))))))).................)))...................... (-17.45 = -17.70 + 0.25)



| Location | 4,664,652 – 4,664,765 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.56 |
| Mean single sequence MFE | -30.30 |
| Consensus MFE | -26.77 |
| Energy contribution | -27.52 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.11 |
| SVM RNA-class probability | 0.988247 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4664652 113 - 22224390 UUUAAGUUCUAUUA------UUCGCCAU-GGAAAUUUAAUUGAAUUAGCCAUUUAGCGUUUUUGGCUAAUUUGCAUGUUGUGUCCAAAUAAAACAUUUGAUGAUGCAGCAUGUGCGUUGC .(((((((((((..------......))-)).)))))))..((((((((((...........))))))))))(((((((((((((((((.....)))))..))))))))))))....... ( -31.20) >DroSec_CAF1 25985 114 - 1 UUUAAGUUUUAUUA------UUCGCCAUUGGAAAUUUAAUUGAAUUAGCCAUUUAGCGUUUUUGGCUAAUUUGCAUGUUGUGUCCAAAUAAAACAUUUGAUGAUGCAGCAUGUGCGUUGC ..............------..(((................((((((((((...........))))))))))(((((((((((((((((.....)))))..))))))))))))))).... ( -30.30) >DroEre_CAF1 26740 113 - 1 UCUAAGUUUUAUUA------UUCGCCAU-AGAAAUUUAAUUGAAUUAGCCAUUUAGCGUUUUUGGCUAAUUUGCAUGUUUUGUCCAAAUAAAACAUUUGAUGAUGCAGCAUGUGCGUUGC ..............------..(((...-............((((((((((...........))))))))))(((((((.(((((((((.....)))))..)))).)))))))))).... ( -24.00) >DroYak_CAF1 23150 119 - 1 UUUAAGUUUCAUUGUGGAAACUCGCAAU-GGAAAUUUAAUUGAAUUAGCCAUUUAGCGUUUUUGGCUAAUUUGCAUGUUUUGUCGAAAUAAAACAUUUGAUGAUGCAGCAUGUGCGCUGC .((((((((((((((((....)))))))-.)))))))))..((((((((((...........))))))))))..(((((((((....)))))))))........(((((......))))) ( -35.70) >consensus UUUAAGUUUUAUUA______UUCGCCAU_GGAAAUUUAAUUGAAUUAGCCAUUUAGCGUUUUUGGCUAAUUUGCAUGUUGUGUCCAAAUAAAACAUUUGAUGAUGCAGCAUGUGCGUUGC ......................(((................((((((((((...........))))))))))(((((((((((((((((.....)))))..))))))))))))))).... (-26.77 = -27.52 + 0.75)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:15:44 2006