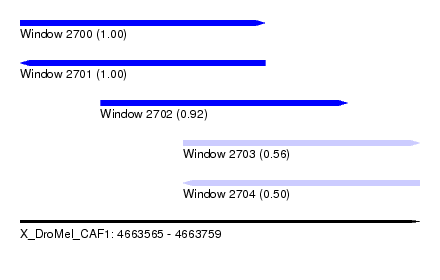
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,663,565 – 4,663,759 |
| Length | 194 |
| Max. P | 0.999970 |

| Location | 4,663,565 – 4,663,684 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.31 |
| Mean single sequence MFE | -31.85 |
| Consensus MFE | -19.33 |
| Energy contribution | -20.45 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.25 |
| Structure conservation index | 0.61 |
| SVM decision value | 4.21 |
| SVM RNA-class probability | 0.999837 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4663565 119 + 22224390 -UUGAGGGAAAUGGGAAAAGCAUAUUAUAAUGUACUUUAGGACGCUUUACAAAGAAAAAGGGAAUCACUUUAAAUAAAGUACACUUUUCCCAUCUGGGAGAAUCCUUUUCAGCAAAUUUG -(((..((((((((((((((..........(((((((((.....(((....)))...((((......))))...)))))))))))))))))))..(((.....)))))))..)))..... ( -25.90) >DroSec_CAF1 24898 120 + 1 GUUCGCGUGAAUGGGAAAAGCUUUCUAUAAAGUACUUUAGGACGCUUCAUGCAGCAAAAGGGAACCACUUUAAAUAAAGUACACUUUUCCCAUCUGGGAGAAUCCUUUUCAGCGAAUUUG ((((((.((((.((((...............((((((((....(((......))).(((((....).))))...))))))))..(((((((....))))))))))).))))))))))... ( -36.30) >DroEre_CAF1 25686 117 + 1 GAUCGCAUGGCUGGGAAAAGCUUUU-UUAAUGUACUUUAGAACUCUUUACGUUG--AAAGGAAAUCACUUUAAAUAAAGUACACUUUUCCCAGCUGGGAGAAUCCUUUUCAGCGAAUUUG ..((((..((((((((((((.((((-((((((((....((....)).)))))))--))))).....(((((....)))))...))))))))))))(((.....))).....))))..... ( -36.90) >DroYak_CAF1 22076 93 + 1 GAUCGCUUGAAUGGGAAAAGCUUUU------------------------CGUUU--AAA-UAAAUCACUUUAAAUAAAGUACACUUUUCCCAUCGGGGAGAAUCCCUUUCAGCGAAUUUG ..(((((.(((.((((...(((((.------------------------.((((--(((-........))))))))))))....(((((((....))))))))))).))))))))..... ( -28.30) >consensus GAUCGCGUGAAUGGGAAAAGCUUUU_AUAAUGUACUUUAGGACGCUUUACGUAG__AAAGGAAAUCACUUUAAAUAAAGUACACUUUUCCCAUCUGGGAGAAUCCUUUUCAGCGAAUUUG ..((((.((((.((((..................................................(((((....)))))....(((((((....))))))))))).))))))))..... (-19.33 = -20.45 + 1.12)

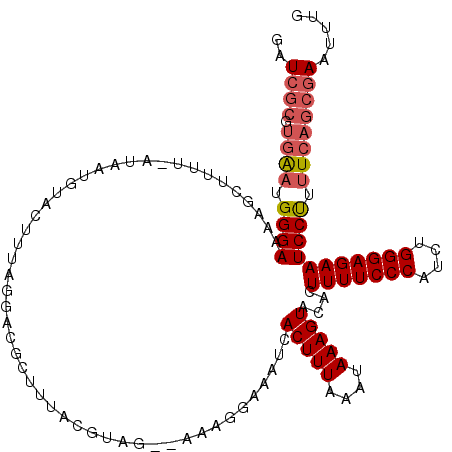
| Location | 4,663,565 – 4,663,684 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.31 |
| Mean single sequence MFE | -30.60 |
| Consensus MFE | -21.96 |
| Energy contribution | -22.40 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.45 |
| Structure conservation index | 0.72 |
| SVM decision value | 5.05 |
| SVM RNA-class probability | 0.999970 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4663565 119 - 22224390 CAAAUUUGCUGAAAAGGAUUCUCCCAGAUGGGAAAAGUGUACUUUAUUUAAAGUGAUUCCCUUUUUCUUUGUAAAGCGUCCUAAAGUACAUUAUAAUAUGCUUUUCCCAUUUCCCUCAA- .........(((...((......))((((((((((((((((((((((..((((.((.......)).)))))))))).((.(....).)).......))))))))))))))))...))).- ( -23.60) >DroSec_CAF1 24898 120 - 1 CAAAUUCGCUGAAAAGGAUUCUCCCAGAUGGGAAAAGUGUACUUUAUUUAAAGUGGUUCCCUUUUGCUGCAUGAAGCGUCCUAAAGUACUUUAUAGAAAGCUUUUCCCAUUCACGCGAAC ....(((((((((..((......))...((((((((((...((.....(((((((.((......((((......)))).....)).))))))).))...)))))))))))))).))))). ( -34.50) >DroEre_CAF1 25686 117 - 1 CAAAUUCGCUGAAAAGGAUUCUCCCAGCUGGGAAAAGUGUACUUUAUUUAAAGUGAUUUCCUUU--CAACGUAAAGAGUUCUAAAGUACAUUAA-AAAAGCUUUUCCCAGCCAUGCGAUC .....((((((....((......)).((((((((((((((((((((((((...(((.......)--))...))))......)))))))).....-....)))))))))))))).)))).. ( -33.70) >DroYak_CAF1 22076 93 - 1 CAAAUUCGCUGAAAGGGAUUCUCCCCGAUGGGAAAAGUGUACUUUAUUUAAAGUGAUUUA-UUU--AAACG------------------------AAAAGCUUUUCCCAUUCAAGCGAUC .....(((((....(((......)))((((((((((((....((((..((((....))))-..)--)))..------------------------....))))))))))))..))))).. ( -30.60) >consensus CAAAUUCGCUGAAAAGGAUUCUCCCAGAUGGGAAAAGUGUACUUUAUUUAAAGUGAUUCCCUUU__CAACGUAAAGCGUCCUAAAGUACAUUAU_AAAAGCUUUUCCCAUUCACGCGAAC .....((((......((......)).((((((((((((.((((((....)))))).....((((........)))).......................))))))))))))...)))).. (-21.96 = -22.40 + 0.44)

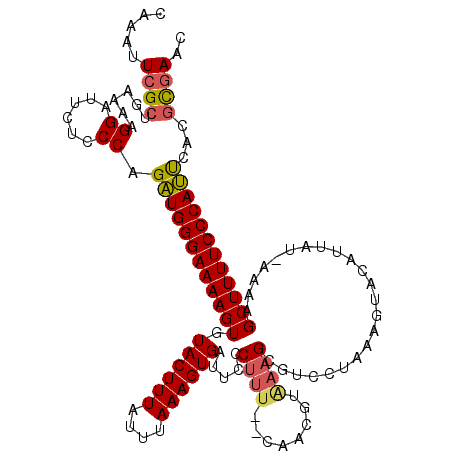
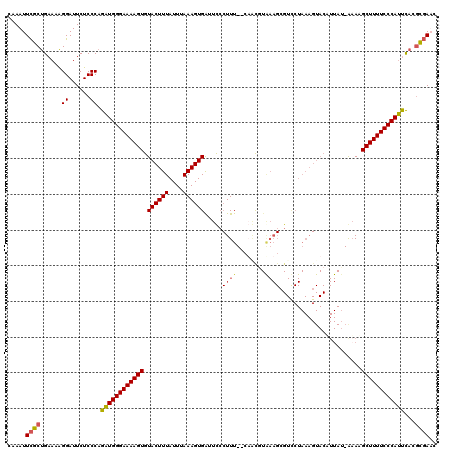
| Location | 4,663,604 – 4,663,724 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.33 |
| Mean single sequence MFE | -25.75 |
| Consensus MFE | -18.33 |
| Energy contribution | -17.95 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.918598 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4663604 120 + 22224390 GACGCUUUACAAAGAAAAAGGGAAUCACUUUAAAUAAAGUACACUUUUCCCAUCUGGGAGAAUCCUUUUCAGCAAAUUUGCGUUAAAAUAAACAGCAAGAAAAGUACAUCAAAAGGGUAA .((.((((.....(..(((((((...(((((....)))))....(((((((....))))))))))))))...)...((((((((......))).)))))............)))).)).. ( -24.60) >DroSec_CAF1 24938 120 + 1 GACGCUUCAUGCAGCAAAAGGGAACCACUUUAAAUAAAGUACACUUUUCCCAUCUGGGAGAAUCCUUUUCAGCGAAUUUGUGUUAAAAUAAACAGCAAGAAAAGUACAUCAAAAGGGUAA .((..(((.(((.((.(((((((...(((((....)))))....(((((((....))))))))))))))..)).......((((......))))))).)))..))............... ( -26.00) >DroEre_CAF1 25725 118 + 1 AACUCUUUACGUUG--AAAGGAAAUCACUUUAAAUAAAGUACACUUUUCCCAGCUGGGAGAAUCCUUUUCAGCGAAUUUGCGUUAAAAUAAACAGCAAGAAAAGUACGUCAAAAGGGUAA .(((((((.(((((--((((((....(((((....)))))....(((((((....)))))))))).))))))))..((((((((......))).)))))............))))))).. ( -33.60) >DroYak_CAF1 22101 108 + 1 ---------CGUUU--AAA-UAAAUCACUUUAAAUAAAGUACACUUUUCCCAUCGGGGAGAAUCCCUUUCAGCGAAUUUGCGUUAAAAUAAACAGCAAGAAAAGUACAUCAAAAGGGUAA ---------.((((--(((-........))))))).........(((((((....))))))).((((((..((...((((((((......))).)))))....))......))))))... ( -18.80) >consensus GACGCUUUACGUAG__AAAGGAAAUCACUUUAAAUAAAGUACACUUUUCCCAUCUGGGAGAAUCCUUUUCAGCGAAUUUGCGUUAAAAUAAACAGCAAGAAAAGUACAUCAAAAGGGUAA .......................(((.((((.......((((..(((((((....)))))))..............((((((((......))).)))))....))))....))))))).. (-18.33 = -17.95 + -0.38)

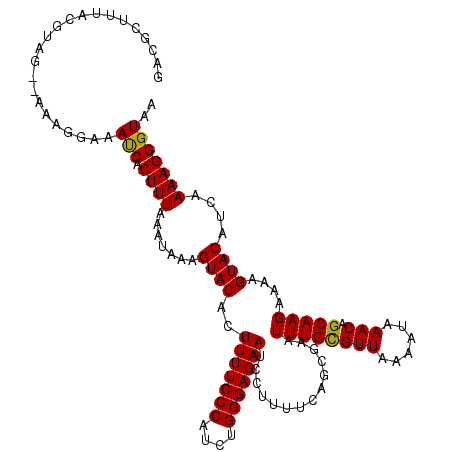
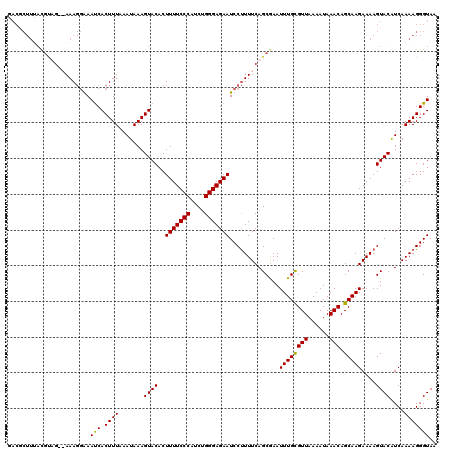
| Location | 4,663,644 – 4,663,759 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.20 |
| Mean single sequence MFE | -23.88 |
| Consensus MFE | -17.46 |
| Energy contribution | -17.77 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.558503 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4663644 115 + 22224390 ACACUUUUCCCAUCUGGGAGAAUCCUUUUCAGCAAAUUUGCGUUAAAAUAAACAGCAAGAAAAGUACAUCAAAAGGGUAAAGUGAACA-----AAGAAUACCAUGGGAAUAUUCCUUGAG .((((((.(((..(((..((.....))..)))....((((((((......))).)))))...............))).))))))..((-----(.(((((((...))..))))).))).. ( -24.20) >DroSec_CAF1 24978 115 + 1 ACACUUUUCCCAUCUGGGAGAAUCCUUUUCAGCGAAUUUGUGUUAAAAUAAACAGCAAGAAAAGUACAUCAAAAGGGUAAAGUGAAAA-----AAGAAUACCAUGGGAAUAUUCCUUGAG .((((((((((....))))))((((((((..((...(((((......)))))..))..((........))))))))))..))))....-----........((.((((....)))))).. ( -23.10) >DroEre_CAF1 25763 113 + 1 ACACUUUUCCCAGCUGGGAGAAUCCUUUUCAGCGAAUUUGCGUUAAAAUAAACAGCAAGAAAAGUACGUCAAAAGGGUAAAA-------GGGAAAAAACACCAUGGGAAUAUUCCUUGAG ....(((((((.((((..((.....))..))))...((((((((......))).))))).......................-------))))))).....((.((((....)))))).. ( -25.10) >DroYak_CAF1 22129 120 + 1 ACACUUUUCCCAUCGGGGAGAAUCCCUUUCAGCGAAUUUGCGUUAAAAUAAACAGCAAGAAAAGUACAUCAAAAGGGUAAAAAAAAAAUGGGGAAAAACACCAUGGGAAUAUUGCUUGAG ......(((((((.((......(((((.........((((((((......))).))))).....(((.((....)))))..........)))))......)))))))))........... ( -23.10) >consensus ACACUUUUCCCAUCUGGGAGAAUCCUUUUCAGCGAAUUUGCGUUAAAAUAAACAGCAAGAAAAGUACAUCAAAAGGGUAAAAUGAAAA_____AAAAACACCAUGGGAAUAUUCCUUGAG .....((((((....))))))((((((((..((...((((((((......))).)))))....)).....)))))))).......................((.((((....)))))).. (-17.46 = -17.77 + 0.31)

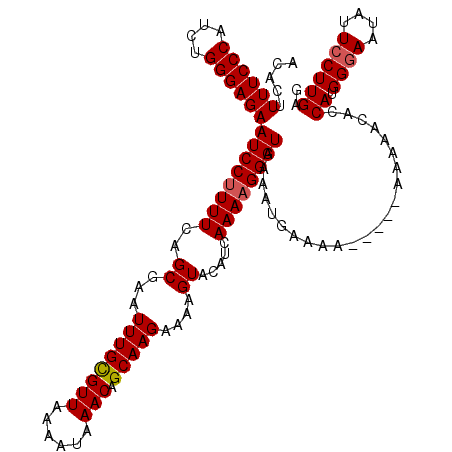

| Location | 4,663,644 – 4,663,759 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.20 |
| Mean single sequence MFE | -24.65 |
| Consensus MFE | -17.02 |
| Energy contribution | -16.64 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.69 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4663644 115 - 22224390 CUCAAGGAAUAUUCCCAUGGUAUUCUU-----UGUUCACUUUACCCUUUUGAUGUACUUUUCUUGCUGUUUAUUUUAACGCAAAUUUGCUGAAAAGGAUUCUCCCAGAUGGGAAAAGUGU ..(((((((((((.....)))))))))-----))..((((((..((((((.(.(((......((((.(((......)))))))...)))).))))))....((((....)))))))))). ( -28.70) >DroSec_CAF1 24978 115 - 1 CUCAAGGAAUAUUCCCAUGGUAUUCUU-----UUUUCACUUUACCCUUUUGAUGUACUUUUCUUGCUGUUUAUUUUAACACAAAUUCGCUGAAAAGGAUUCUCCCAGAUGGGAAAAGUGU ...((((((((((.....)))))))))-----)...((((((.(((.((((..(..((((((..((((((......)))).......)).))))))....)...)))).))).)))))). ( -24.91) >DroEre_CAF1 25763 113 - 1 CUCAAGGAAUAUUCCCAUGGUGUUUUUUCCC-------UUUUACCCUUUUGACGUACUUUUCUUGCUGUUUAUUUUAACGCAAAUUCGCUGAAAAGGAUUCUCCCAGCUGGGAAAAGUGU .(((((((....)))...((((.........-------...))))...))))..((((((((((((((....(((((.((......)).))))).((.....)))))).)))))))))). ( -21.20) >DroYak_CAF1 22129 120 - 1 CUCAAGCAAUAUUCCCAUGGUGUUUUUCCCCAUUUUUUUUUUACCCUUUUGAUGUACUUUUCUUGCUGUUUAUUUUAACGCAAAUUCGCUGAAAGGGAUUCUCCCCGAUGGGAAAAGUGU .....(((...(((((((((.(.....).))............(((((((..((........((((.(((......)))))))...))..)))))))..........)))))))...))) ( -23.80) >consensus CUCAAGGAAUAUUCCCAUGGUAUUCUU_____UUUUCACUUUACCCUUUUGAUGUACUUUUCUUGCUGUUUAUUUUAACGCAAAUUCGCUGAAAAGGAUUCUCCCAGAUGGGAAAAGUGU ...........(((((((((((...................)))).....((....((((((..((((((......)))).......)).))))))...))......)))))))...... (-17.02 = -16.64 + -0.38)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:15:38 2006