
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,663,120 – 4,663,336 |
| Length | 216 |
| Max. P | 0.960653 |

| Location | 4,663,120 – 4,663,220 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 88.61 |
| Mean single sequence MFE | -26.30 |
| Consensus MFE | -19.61 |
| Energy contribution | -19.67 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.823079 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4663120 100 + 22224390 AGUUCAUCAGCUUACUGUUCGCCA--GACCCCCGAAGGCCUUUCCAAGCCACCUUUUGGAAUGGGCCCAGUUCAUUGCCGAAUAAUGAACAUAACAAAUCAA .(((((((.((..((((((((...--......))))((((((((((((......))))))).))))))))).....)).))....)))))............ ( -25.40) >DroSec_CAF1 24448 99 + 1 AGUUCUUCAGCUUACUGUUCGCCA--GAACCCCGAAGGCCUUUCCAAGCCAACC-CUGGAAUGGGGCCACUUCAUUGCCGAAUAAUGAACAUAACAAAUCAA .(((((((.((.....((((....--))))...(((((((...(((..(((...-.)))..)))))))..)))...)).)))....))))............ ( -22.90) >DroEre_CAF1 25235 100 + 1 AGUUCAUCAGCUUUCUGUUCGCCA--GACCCCCGAAGGCCUUUCCAUGCCACCCCGUGGAAUGGGGCCAGUUCAUUGCCGAAUAAUGAACAUAACAAAUCAA .............((((.....))--)).....((.((((((((((((......)))))))..))))).((((((((.....))))))))........)).. ( -26.60) >DroYak_CAF1 21590 101 + 1 AGUUCAUCAGCUUACUGUUCGGCGAAAGCCCCCAAAGGUCUUGCCAUGCCACCCCA-GGAAUGGGGCCUGUUCAUUGCCGAAUAAUGAACAUAACAAAUCAA .((((((........((((((((((.((.(((((..((.....))...((......-))..))))).)).....))))))))))))))))............ ( -30.30) >consensus AGUUCAUCAGCUUACUGUUCGCCA__GACCCCCGAAGGCCUUUCCAAGCCACCCCAUGGAAUGGGGCCAGUUCAUUGCCGAAUAAUGAACAUAACAAAUCAA .((((((........((((((.((..((((((((..(((........))).((....))..)))))...)))...)).))))))))))))............ (-19.61 = -19.67 + 0.06)


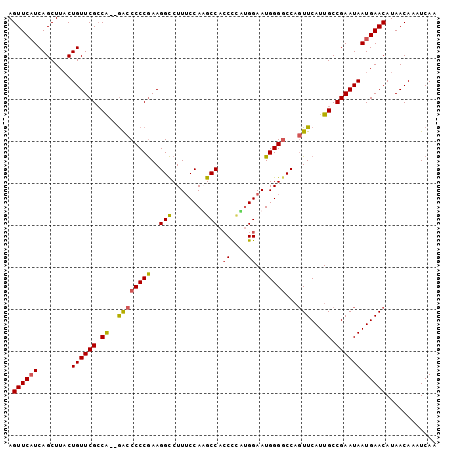
| Location | 4,663,142 – 4,663,260 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.78 |
| Mean single sequence MFE | -41.70 |
| Consensus MFE | -30.77 |
| Energy contribution | -33.52 |
| Covariance contribution | 2.75 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.97 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.960653 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4663142 118 + 22224390 CA--GACCCCCGAAGGCCUUUCCAAGCCACCUUUUGGAAUGGGCCCAGUUCAUUGCCGAAUAAUGAACAUAACAAAUCAACGGCUCGUCAUUAGGCGAGCCCCAGCAAAAUGGGGCUAGG .(--(.((((.((.((((((((((((......))))))).)))))..((((((((.....))))))))........))...((((((((....))))))))..........))))))... ( -45.20) >DroSec_CAF1 24470 117 + 1 CA--GAACCCCGAAGGCCUUUCCAAGCCAACC-CUGGAAUGGGGCCACUUCAUUGCCGAAUAAUGAACAUAACAAAUCAACGGCUCGUCAUUAGGCGAGCCCCAGUAAAAUGGGGCUAGG ..--...((((((.((((...(((..(((...-.)))..)))))))..(((((((.....))))))).........))...((((((((....))))))))..........))))..... ( -39.60) >DroEre_CAF1 25257 114 + 1 CA--GACCCCCGAAGGCCUUUCCAUGCCACCCCGUGGAAUGGGGCCAGUUCAUUGCCGAAUAAUGAACAUAACAAAUCAACGGCUCGUCAUUAGGCC----CCGGUAAAAUGGGGCUAGG ..--(((..(((..((((((((((((......)))))))..))))).((((((((.....))))))))............)))...)))....((((----(((......)))))))... ( -43.70) >DroYak_CAF1 21612 117 + 1 CGAAAGCCCCCAAAGGUCUUGCCAUGCCACCCCA-GGAAUGGGGCCUGUUCAUUGCCGAAUAAUGAACAUAACAAAUCAACGGCUCGUCAUUAGGCC--CCCCAGUGAAAUGGGGCUAGG ....((((((....((.....))..(((.(((((-....)))))..(((((((((.....)))))))))............)))...(((((.((..--..)))))))...))))))... ( -38.30) >consensus CA__GACCCCCGAAGGCCUUUCCAAGCCACCCCAUGGAAUGGGGCCAGUUCAUUGCCGAAUAAUGAACAUAACAAAUCAACGGCUCGUCAUUAGGCC__CCCCAGUAAAAUGGGGCUAGG .......(((((..((((((((((((......)))))))..))))).((((((((.....)))))))).............((((((((....)))))))).........)))))..... (-30.77 = -33.52 + 2.75)


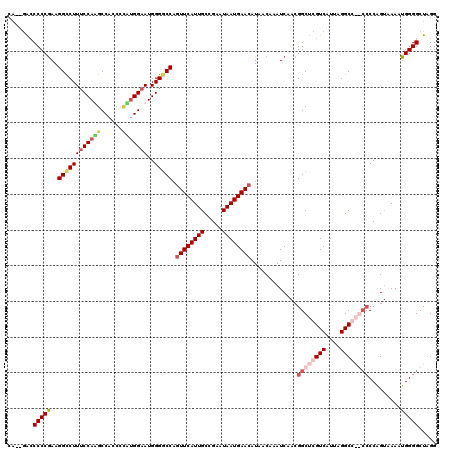
| Location | 4,663,180 – 4,663,296 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.13 |
| Mean single sequence MFE | -29.55 |
| Consensus MFE | -20.34 |
| Energy contribution | -22.65 |
| Covariance contribution | 2.31 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.917899 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4663180 116 - 22224390 CCCAUUCCCAUUCCCAUUCCCAUUCCCG----UUCGCAUUCCUAGCCCCAUUUUGCUGGGGCUCGCCUAAUGACGAGCCGUUGAUUUGUUAUGUUCAUUAUUCGGCAAUGAACUGGGCCC ........................((((----((((.......(((((((......))))))).(((.(((((.((((...(((....))).)))).))))).)))..))))).)))... ( -31.40) >DroSec_CAF1 24507 108 - 1 CCCAUUCCCAUUCGCAUUUC------------UUCGCAUUCCUAGCCCCAUUUUACUGGGGCUCGCCUAAUGACGAGCCGUUGAUUUGUUAUGUUCAUUAUUCGGCAAUGAAGUGGCCCC ..............((((((------------.(((((((.(.(((((((......))))))).)...)))).)))((((.(((..........))).....))))...))))))..... ( -29.20) >DroEre_CAF1 25295 116 - 1 CCCAUUCCCAUUCCCAUUCCAGUUCCCAUUUCUUCGCAUUCCUAGCCCCAUUUUACCGG----GGCCUAAUGACGAGCCGUUGAUUUGUUAUGUUCAUUAUUCGGCAAUGAACUGGCCCC ..................(((((((........(((((((....(((((........))----)))..)))).)))((((.(((..........))).....))))...))))))).... ( -31.00) >DroYak_CAF1 21651 118 - 1 CCCAUUCCCAUUUCUAUUCCCGUUUCCAUUUCUUUGCAUUCCUAGCCCCAUUUCACUGGGG--GGCCUAAUGACGAGCCGUUGAUUUGUUAUGUUCAUUAUUCGGCAAUGAACAGGCCCC ..............................................((((......))))(--(((((.((((((((.......))))))))(((((((.......))))))))))))). ( -26.60) >consensus CCCAUUCCCAUUCCCAUUCCCGUUCCCA____UUCGCAUUCCUAGCCCCAUUUUACUGGGG__CGCCUAAUGACGAGCCGUUGAUUUGUUAUGUUCAUUAUUCGGCAAUGAACUGGCCCC .((((((..........................(((((((.(((((((((......)))))))))...)))).)))((((.(((..........))).....))))...))).))).... (-20.34 = -22.65 + 2.31)


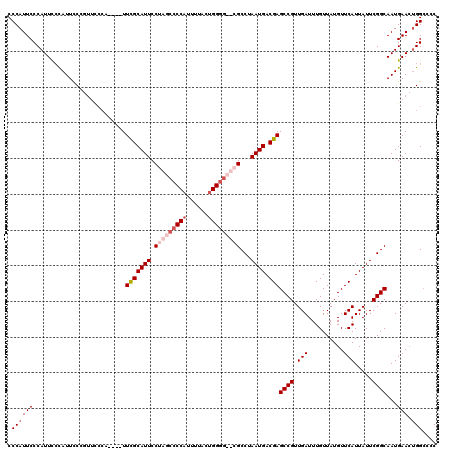
| Location | 4,663,220 – 4,663,336 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.97 |
| Mean single sequence MFE | -31.96 |
| Consensus MFE | -21.25 |
| Energy contribution | -22.88 |
| Covariance contribution | 1.62 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.560764 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4663220 116 + 22224390 CGGCUCGUCAUUAGGCGAGCCCCAGCAAAAUGGGGCUAGGAAUGCGAA----CGGGAAUGGGAAUGGGAAUGGGAAUGGGACCUAUGCAUAAUUUUUCAGCUCACUGAAGCAAAAACAAA .((((((((....))))))))(((((........))).))..(((...----(((((.((..((...(.(((((.......))))).).....))..))..)).)))..)))........ ( -33.00) >DroSec_CAF1 24547 108 + 1 CGGCUCGUCAUUAGGCGAGCCCCAGUAAAAUGGGGCUAGGAAUGCGAA------------GAAAUGCGAAUGGGAAUGGGUCCUAUGCAUAAUUUUUCAGCUCACUGAAGCAAAAACAAA .((((((((....))))))))(((......))).(((.((...(((((------------(((((((..((((((.....))))))))))..)))))).))...))..)))......... ( -35.00) >DroEre_CAF1 25335 116 + 1 CGGCUCGUCAUUAGGCC----CCGGUAAAAUGGGGCUAGGAAUGCGAAGAAAUGGGAACUGGAAUGGGAAUGGGAAUGGGUCCUAUGCAUAAUUUUUCAGCUCACUGAAGCAAAAACAAA ..(((((.((((.((((----(((......)))))))...)))))))......((((.((((((.....((((((.....))))))........)))))).)).))...))......... ( -32.02) >DroYak_CAF1 21691 118 + 1 CGGCUCGUCAUUAGGCC--CCCCAGUGAAAUGGGGCUAGGAAUGCAAAGAAAUGGAAACGGGAAUAGAAAUGGGAAUGGGUCCUAUGCAUAAUUUUUCAGCUCACUGAAGCAAAAACAAA .((((........))))--(((((......)))))......((((.......((....)).........((((((.....))))))))))....((((((....)))))).......... ( -27.80) >consensus CGGCUCGUCAUUAGGCC__CCCCAGUAAAAUGGGGCUAGGAAUGCGAA____UGGGAACGGGAAUGGGAAUGGGAAUGGGUCCUAUGCAUAAUUUUUCAGCUCACUGAAGCAAAAACAAA ..(((((.((((.....(((((((......)))))))...)))))))..............((((....((((((.....)))))).....))))(((((....)))))))......... (-21.25 = -22.88 + 1.62)

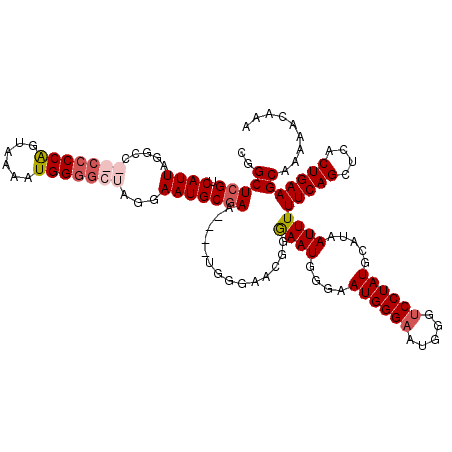
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:15:34 2006