
| Sequence ID | X_DroMel_CAF1 |
|---|---|
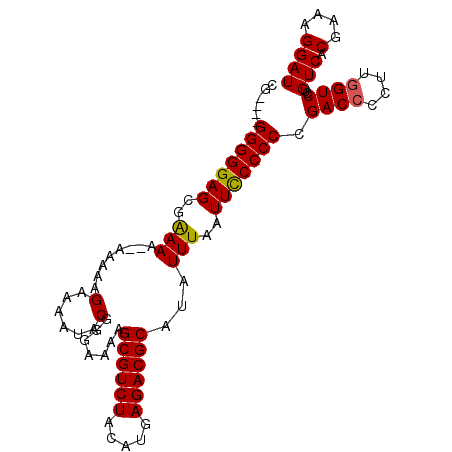
| Location | 4,660,072 – 4,660,173 |
| Length | 101 |
| Max. P | 0.882081 |

| Location | 4,660,072 – 4,660,173 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 93.22 |
| Mean single sequence MFE | -32.12 |
| Consensus MFE | -25.20 |
| Energy contribution | -25.07 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.45 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.882081 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4660072 101 + 22224390 CGAAGGGGGGGAGCGGAAAAAAAAAAGAAAAUACGGGAAAAGCGUCUACAUGAGACGCAUAUUUAAUUUCCCCCGACCCCUUGGUCCUGUCACGAAAGGAU ....((((((...(((..........(......)((((((.((((((.....))))))........))))))))).)))))).(((((........))))) ( -30.80) >DroSec_CAF1 21477 94 + 1 CG----GGGGGAGCGAAAA---AAAAGAAAAUACGGGAAAAGCGUCUACAUGAGACGCAUAUUUAAUUCCCCCCGACCCCUUGGUCCUGUCACGAAAGGAU .(----(((((((..(((.---....(......).......((((((.....))))))...)))..))))))))((((....))))..(((.(....)))) ( -31.60) >DroSim_CAF1 10425 95 + 1 CG----GGGGGAGCGAAAA--AAAAAGAAAAUACGGGAAAAGCGUCUACAUGAGACGCAUAUUUAAUUCCCCCCGACCCCUUGGUCCUGUCACGAAAGGAU .(----(((((((..(((.--.....(......).......((((((.....))))))...)))..))))))))((((....))))..(((.(....)))) ( -31.60) >DroEre_CAF1 22316 95 + 1 CG----GGGGGAGUGAAA--AAAAAGGAAAAUACGGGAAAAGCGUCUACAUGAGACGCAUAUUUAAUUCCCCCCGACCCCUUCGUCCUGUCACGAAAGGAU ((----((((((((.(((--.....(.......).......((((((.....))))))...))).))))))))))..((.(((((......))))).)).. ( -34.50) >consensus CG____GGGGGAGCGAAAA__AAAAAGAAAAUACGGGAAAAGCGUCUACAUGAGACGCAUAUUUAAUUCCCCCCGACCCCUUGGUCCUGUCACGAAAGGAU ......(((((((..(((........(......).......((((((.....))))))...)))..))))))).((((....))))..(((.(....)))) (-25.20 = -25.07 + -0.13)


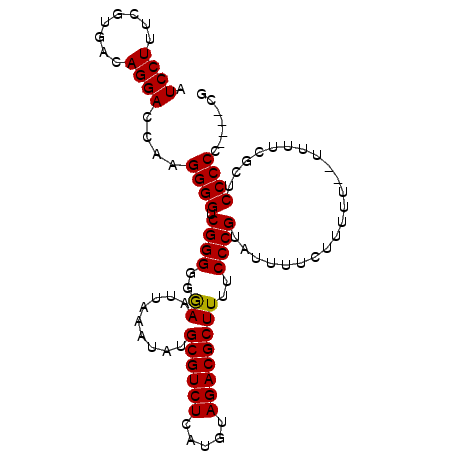
| Location | 4,660,072 – 4,660,173 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 93.22 |
| Mean single sequence MFE | -29.94 |
| Consensus MFE | -23.99 |
| Energy contribution | -23.80 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.875576 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4660072 101 - 22224390 AUCCUUUCGUGACAGGACCAAGGGGUCGGGGGAAAUUAAAUAUGCGUCUCAUGUAGACGCUUUUCCCGUAUUUUCUUUUUUUUUUCCGCUCCCCCCCUUCG .((((........))))..((((((..((((((((..(((...((((((.....)))))).......(......)..)))..))))).)))..)))))).. ( -28.10) >DroSec_CAF1 21477 94 - 1 AUCCUUUCGUGACAGGACCAAGGGGUCGGGGGGAAUUAAAUAUGCGUCUCAUGUAGACGCUUUUCCCGUAUUUUCUUUU---UUUUCGCUCCCCC----CG (((((((.((......)).)))))))((((((((.........((((((.....))))))......((...........---....)).))))))----)) ( -28.76) >DroSim_CAF1 10425 95 - 1 AUCCUUUCGUGACAGGACCAAGGGGUCGGGGGGAAUUAAAUAUGCGUCUCAUGUAGACGCUUUUCCCGUAUUUUCUUUUU--UUUUCGCUCCCCC----CG (((((((.((......)).)))))))((((((((.........((((((.....)))))).......(......).....--.......))))))----)) ( -28.70) >DroEre_CAF1 22316 95 - 1 AUCCUUUCGUGACAGGACGAAGGGGUCGGGGGGAAUUAAAUAUGCGUCUCAUGUAGACGCUUUUCCCGUAUUUUCCUUUUU--UUUCACUCCCCC----CG ((((((((((......))))))))))((((((((.........((((((.....)))))).......(......)......--......))))))----)) ( -34.20) >consensus AUCCUUUCGUGACAGGACCAAGGGGUCGGGGGGAAUUAAAUAUGCGUCUCAUGUAGACGCUUUUCCCGUAUUUUCUUUUU__UUUUCGCUCCCCC____CG .((((........))))....((((.((((..((.........((((((.....))))))))..))))......................))))....... (-23.99 = -23.80 + -0.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:15:28 2006