
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,654,558 – 4,654,874 |
| Length | 316 |
| Max. P | 0.999950 |

| Location | 4,654,558 – 4,654,654 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.47 |
| Mean single sequence MFE | -45.82 |
| Consensus MFE | -13.44 |
| Energy contribution | -14.44 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.29 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.925930 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4654558 96 + 22224390 CCCCA-UCCACCUUCCAGUUCAGGUGUCCCAGCAUAU------ACAUGUUCUUGGUGGCGG-CACUG----------------GAGGUGAUGGAGGCGGUUGCUGCGGAUGGCCACGUUU ..(((-((.((((.(((((...(.(((((((((((..------..))))...))).)))).-)))))----------------)))))))))).(((.(((......))).)))...... ( -35.50) >DroSec_CAF1 16225 109 + 1 CCCCUCACCACCUUCCAGCUCAGGUGUCCCUGCAUAUA---GUACAUGUUCUUGGUGGCGGGCAAGGGG--------CAGGGAGAGGUGGUGUAGGCGGCUGUGGCGGAUGGCCACGUUU ((((((((((((((((.((((...((((((..(....(---(........))..)..).)))))..)))--------)..))).)))))))).))).))..(((((.....))))).... ( -50.00) >DroSim_CAF1 4881 109 + 1 CCCCUCACCACCUUCCAGCUCAGGUGUCCCUGCAUAUA---GUACAUGUUCUUGGUGGCGGGCAAGGGG--------CAGGGAGAGGUGGUGUAGGCGGCUGUGGCGGAUGGCCACGUUU ((((((((((((((((.((((...((((((..(....(---(........))..)..).)))))..)))--------)..))).)))))))).))).))..(((((.....))))).... ( -50.00) >DroEre_CAF1 16819 95 + 1 CCCCU---CACCUUUCAGCUCAGGUGUCCCUGCAUAUACUCGUACAUAUC-UU-GGGGGAGGCACGUGG-------------------GGGUGAGA-GGGCGUGGCGGAUGGCCACGUUU (((((---((((......((((.((((((((.((.(((........))).-.)-).))).))))).)))-------------------))))))).-)))((((((.....))))))... ( -41.50) >DroYak_CAF1 12414 112 + 1 -CCCU---CACCUUCCAGCUCAGGUGUCCCUGCAUAUACUCGUACAUAUUCUUAGGGGGAGUUGCUUGGAUGCUUGGCAGAG---GGCGGGGGAGG-GGGCGUGGCGGAUGGCCACGUUU -((((---(.(((((..((.(((((((((..(((....(((.((........)).)))....)))..))))))))))).)))---)).)))))...-(((((((((.....))))))))) ( -52.10) >consensus CCCCU__CCACCUUCCAGCUCAGGUGUCCCUGCAUAUA___GUACAUGUUCUUGGUGGCGGGCACGGGG________CAG_G_GAGGUGGUGGAGGCGGCUGUGGCGGAUGGCCACGUUU ..((....(((((........)))))..(((((........................)))))...................................)).((((((.....))))))... (-13.44 = -14.44 + 1.00)

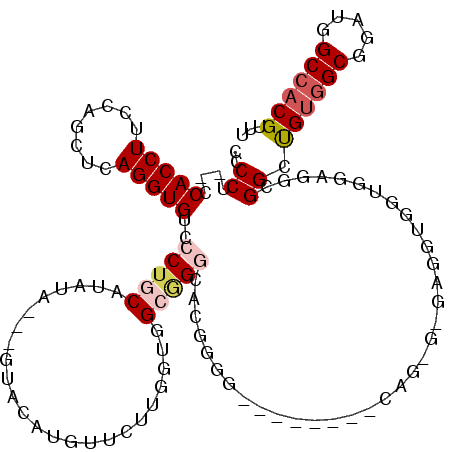
| Location | 4,654,558 – 4,654,654 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.47 |
| Mean single sequence MFE | -36.44 |
| Consensus MFE | -20.45 |
| Energy contribution | -21.54 |
| Covariance contribution | 1.09 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.654400 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4654558 96 - 22224390 AAACGUGGCCAUCCGCAGCAACCGCCUCCAUCACCUC----------------CAGUG-CCGCCACCAAGAACAUGU------AUAUGCUGGGACACCUGAACUGGAAGGUGGA-UGGGG ....((((....)))).((....))((((((((((((----------------((((.-......(((....(((..------..))).))).........))))).)))).))-))))) ( -28.09) >DroSec_CAF1 16225 109 - 1 AAACGUGGCCAUCCGCCACAGCCGCCUACACCACCUCUCCCUG--------CCCCUUGCCCGCCACCAAGAACAUGUAC---UAUAUGCAGGGACACCUGAGCUGGAAGGUGGUGAGGGG ....(((((.....)))))..((.(((.((((((((.((((((--------(..((((........))))...((((..---.)))))))))))..((......)).))))))))))))) ( -41.70) >DroSim_CAF1 4881 109 - 1 AAACGUGGCCAUCCGCCACAGCCGCCUACACCACCUCUCCCUG--------CCCCUUGCCCGCCACCAAGAACAUGUAC---UAUAUGCAGGGACACCUGAGCUGGAAGGUGGUGAGGGG ....(((((.....)))))..((.(((.((((((((.((((((--------(..((((........))))...((((..---.)))))))))))..((......)).))))))))))))) ( -41.70) >DroEre_CAF1 16819 95 - 1 AAACGUGGCCAUCCGCCACGCCC-UCUCACCC-------------------CCACGUGCCUCCCCC-AA-GAUAUGUACGAGUAUAUGCAGGGACACCUGAGCUGAAAGGUG---AGGGG ...((((((.....)))))).((-(((((((.-------------------...(((((.((....-..-))...)))))(((.....((((....)))).)))....))))---))))) ( -35.50) >DroYak_CAF1 12414 112 - 1 AAACGUGGCCAUCCGCCACGCCC-CCUCCCCCGCC---CUCUGCCAAGCAUCCAAGCAACUCCCCCUAAGAAUAUGUACGAGUAUAUGCAGGGACACCUGAGCUGGAAGGUG---AGGG- ...((((((.....))))))..(-((((((.....---...(((...)))((((.((...............((((((....))))))((((....)))).)))))).)).)---))))- ( -35.20) >consensus AAACGUGGCCAUCCGCCACACCCGCCUCCACCACCUC_C_CUG________CCCCGUGCCCGCCACCAAGAACAUGUAC___UAUAUGCAGGGACACCUGAGCUGGAAGGUGG__AGGGG ....(((((.....)))))..((.(((...(((((.....................................((((((....))))))((((....))))........)))))..))))) (-20.45 = -21.54 + 1.09)


| Location | 4,654,594 – 4,654,694 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.94 |
| Mean single sequence MFE | -22.08 |
| Consensus MFE | -16.56 |
| Energy contribution | -17.00 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.793801 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4654594 100 - 22224390 ACUUUAACACUUUGGUGCCACAUUAAGGCUGCAAAUUGUUAAACGUGGCCAUCCGCAGCAACCGCCUCCAUCACCUC----------------CAGUG-CCGCCACCAAGAACAUGU--- ..(((((((.((((..(((.......)))..)))).))))))).(((((........((....))......(((...----------------..)))-..)))))...........--- ( -20.80) >DroSec_CAF1 16263 111 - 1 ACUUUAACACUUUGGUUCCACAUUAAGGCUGCAAAUUGUUAAACGUGGCCAUCCGCCACAGCCGCCUACACCACCUCUCCCUG--------CCCCUUGCCCGCCACCAAGAACAUGUAC- ..........((((((..(......((((.((.....(.....)(((((.....))))).)).))))...............(--------(.....))..)..)))))).........- ( -20.30) >DroSim_CAF1 4919 111 - 1 ACUUUAACACUUUGGUGCCACAUUAAGGCUGCAAAUUGUUAAACGUGGCCAUCCGCCACAGCCGCCUACACCACCUCUCCCUG--------CCCCUUGCCCGCCACCAAGAACAUGUAC- ..........(((((((.(......((((.((.....(.....)(((((.....))))).)).))))...............(--------(.....))..).))))))).........- ( -23.90) >DroEre_CAF1 16856 98 - 1 ACUUUAACACUUUGGUGCCACAUUAAGGCUGCAAAUUGUUAAACGUGGCCAUCCGCCACGCCC-UCUCACCC-------------------CCACGUGCCUCCCCC-AA-GAUAUGUACG ..(((((((.((((..(((.......)))..)))).)))))))((((((.....))))))...-........-------------------...(((((.((....-..-))...))))) ( -24.50) >DroYak_CAF1 12450 116 - 1 ACUUUAACACUUUCGUGCCACAUUAAGGCUGCAAAUUGUUAAACGUGGCCAUCCGCCACGCCC-CCUCCCCCGCC---CUCUGCCAAGCAUCCAAGCAACUCCCCCUAAGAAUAUGUACG ......(((..(((..(((.......)))(((....((((...((((((.....))))))...-........((.---....))..)))).....)))...........)))..)))... ( -20.90) >consensus ACUUUAACACUUUGGUGCCACAUUAAGGCUGCAAAUUGUUAAACGUGGCCAUCCGCCACACCCGCCUCCACCACCUC_C_CUG________CCCCGUGCCCGCCACCAAGAACAUGUAC_ ..(((((((.((((..(((.......)))..)))).))))))).(((((.....)))))............................................................. (-16.56 = -17.00 + 0.44)

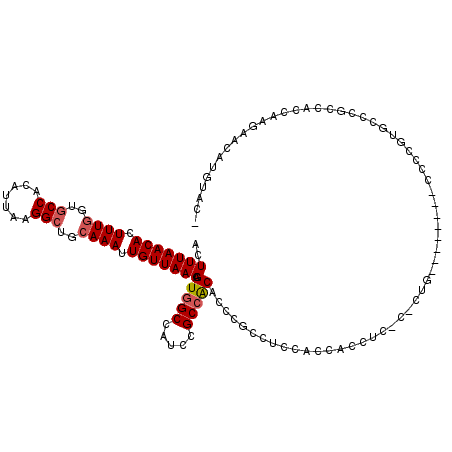

| Location | 4,654,617 – 4,654,734 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.02 |
| Mean single sequence MFE | -42.42 |
| Consensus MFE | -35.06 |
| Energy contribution | -36.18 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.21 |
| SVM RNA-class probability | 0.990284 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4654617 117 + 22224390 ---GAGGUGAUGGAGGCGGUUGCUGCGGAUGGCCACGUUUAACAAUUUGCAGCCUUAAUGUGGCACCAAAGUGUUAAAGUCCAUGUGGCCCAUAAGUUGGCCGGUGGAAAUGGCCGUAAU ---(.((..((.(...).))..)).)..(((((((..(((((((.((((..(((.......)))..)))).))))))).(((((.(((((........))))))))))..)))))))... ( -39.60) >DroSec_CAF1 16294 120 + 1 GGAGAGGUGGUGUAGGCGGCUGUGGCGGAUGGCCACGUUUAACAAUUUGCAGCCUUAAUGUGGAACCAAAGUGUUAAAGUCCAUGUGGCCCAUAAGUUGGCCGGUGGAAAUGGCCGUAAU ...............(((((((((((.....))))).(((((((.((((...((.......))...)))).))))))).(((((.(((((........))))))))))...))))))... ( -40.90) >DroSim_CAF1 4950 120 + 1 GGAGAGGUGGUGUAGGCGGCUGUGGCGGAUGGCCACGUUUAACAAUUUGCAGCCUUAAUGUGGCACCAAAGUGUUAAAGUCCAUGUGGCCCAUAAGUUGGCCGGUGGAAAUGGCCGUAAU ...............(((((((((((.....))))).(((((((.((((..(((.......)))..)))).))))))).(((((.(((((........))))))))))...))))))... ( -43.50) >DroEre_CAF1 16883 111 + 1 --------GGGUGAGA-GGGCGUGGCGGAUGGCCACGUUUAACAAUUUGCAGCCUUAAUGUGGCACCAAAGUGUUAAAGUCCAUGUGGCCCAUAAGUUGGCCGGUGGAAAUGGCCGUAAU --------((((.(.(-(((((((((.....))))).(((((((.((((..(((.......)))..)))).))))))))))).).).)))).......(((((.......)))))..... ( -43.70) >DroYak_CAF1 12490 116 + 1 AG---GGCGGGGGAGG-GGGCGUGGCGGAUGGCCACGUUUAACAAUUUGCAGCCUUAAUGUGGCACGAAAGUGUUAAAGUCCAUGUGGCCCAUAAGUUGGCCGGUGGAAAUGGCCGUAAU ..---.(((((.....-(((((((((.....))))))))).....))))).(((......((((((....))))))...(((((.(((((........))))))))))...)))...... ( -44.40) >consensus _G_GAGGUGGUGGAGGCGGCUGUGGCGGAUGGCCACGUUUAACAAUUUGCAGCCUUAAUGUGGCACCAAAGUGUUAAAGUCCAUGUGGCCCAUAAGUUGGCCGGUGGAAAUGGCCGUAAU ...............(((((((((((.....))))).(((((((.((((..(((.......)))..)))).))))))).(((((.(((((........))))))))))...))))))... (-35.06 = -36.18 + 1.12)

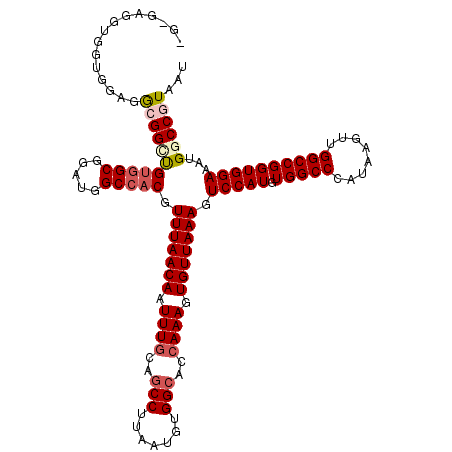

| Location | 4,654,617 – 4,654,734 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.02 |
| Mean single sequence MFE | -34.56 |
| Consensus MFE | -30.92 |
| Energy contribution | -31.32 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.44 |
| Structure conservation index | 0.89 |
| SVM decision value | 4.79 |
| SVM RNA-class probability | 0.999950 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4654617 117 - 22224390 AUUACGGCCAUUUCCACCGGCCAACUUAUGGGCCACAUGGACUUUAACACUUUGGUGCCACAUUAAGGCUGCAAAUUGUUAAACGUGGCCAUCCGCAGCAACCGCCUCCAUCACCUC--- .....((((((.((((..((((........))))...)))).(((((((.((((..(((.......)))..)))).))))))).)))))).......((....))............--- ( -33.60) >DroSec_CAF1 16294 120 - 1 AUUACGGCCAUUUCCACCGGCCAACUUAUGGGCCACAUGGACUUUAACACUUUGGUUCCACAUUAAGGCUGCAAAUUGUUAAACGUGGCCAUCCGCCACAGCCGCCUACACCACCUCUCC ....((((....((((..((((........))))...)))).(((((((.((((...((.......))...)))).))))))).(((((.....))))).))))................ ( -32.60) >DroSim_CAF1 4950 120 - 1 AUUACGGCCAUUUCCACCGGCCAACUUAUGGGCCACAUGGACUUUAACACUUUGGUGCCACAUUAAGGCUGCAAAUUGUUAAACGUGGCCAUCCGCCACAGCCGCCUACACCACCUCUCC ....((((....((((..((((........))))...)))).(((((((.((((..(((.......)))..)))).))))))).(((((.....))))).))))................ ( -36.80) >DroEre_CAF1 16883 111 - 1 AUUACGGCCAUUUCCACCGGCCAACUUAUGGGCCACAUGGACUUUAACACUUUGGUGCCACAUUAAGGCUGCAAAUUGUUAAACGUGGCCAUCCGCCACGCCC-UCUCACCC-------- .....((((.........)))).......((((.........(((((((.((((..(((.......)))..)))).))))))).(((((.....)))))))))-........-------- ( -34.80) >DroYak_CAF1 12490 116 - 1 AUUACGGCCAUUUCCACCGGCCAACUUAUGGGCCACAUGGACUUUAACACUUUCGUGCCACAUUAAGGCUGCAAAUUGUUAAACGUGGCCAUCCGCCACGCCC-CCUCCCCCGCC---CU .....((((.........)))).......((((.....(((.(((((((.(((.(((((.......))).))))).)))))))((((((.....))))))...-..)))...)))---). ( -35.00) >consensus AUUACGGCCAUUUCCACCGGCCAACUUAUGGGCCACAUGGACUUUAACACUUUGGUGCCACAUUAAGGCUGCAAAUUGUUAAACGUGGCCAUCCGCCACACCCGCCUCCACCACCUC_C_ .....((((((.((((..((((........))))...)))).(((((((.((((..(((.......)))..)))).))))))).)))))).............................. (-30.92 = -31.32 + 0.40)

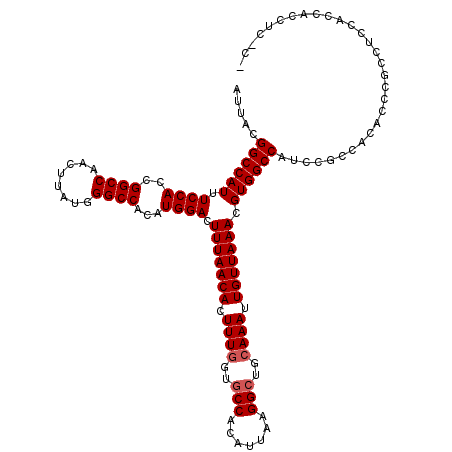
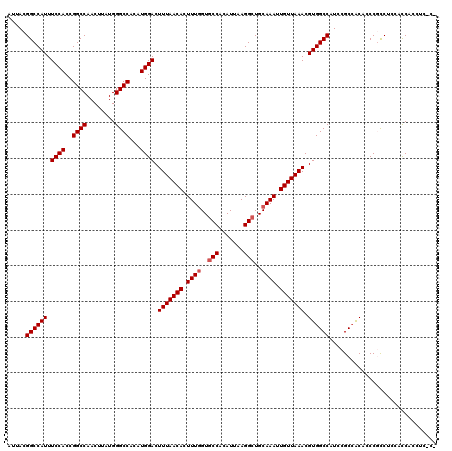
| Location | 4,654,694 – 4,654,794 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.93 |
| Mean single sequence MFE | -28.62 |
| Consensus MFE | -25.36 |
| Energy contribution | -25.56 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.836690 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4654694 100 - 22224390 UCAACCCC-----UUUUCUUCGUGGCCAUUCA---------------AAAGGGAUUGGGCGACACGUACUCCGUCCAUACAUUACGGCCAUUUCCACCGGCCAACUUAUGGGCCACAUGG ........-----........((((((...((---------------.((((...((((((..........))))))..).....((((.........))))..))).)))))))).... ( -28.10) >DroSec_CAF1 16374 100 - 1 UCAACCCC-----UUUUCUUCAUGGCCAUUCA---------------AAAGGGAUUGGGCGACACGUACUCCGUCCAUACAUUACGGCCAUUUCCACCGGCCAACUUAUGGGCCACAUGG ........-----........((((((((((.---------------....))))((((((..........))))))........))))))..(((..((((........))))...))) ( -25.50) >DroSim_CAF1 5030 100 - 1 UCAACCCC-----UUUUCUUCGUGGCCAUUCA---------------AAAGGGAUUGGGCGACACGUACUCCGUCCAUACAUUACGGCCAUUUCCACCGGCCAACUUAUGGGCCACAUGG ........-----........((((((...((---------------.((((...((((((..........))))))..).....((((.........))))..))).)))))))).... ( -28.10) >DroEre_CAF1 16954 99 - 1 UCAACCCC-----UUUUCUUCGUGGCCAUUCA----------------AAGGGAUUGGGCGACACGUACUCGGUCCAUACAUUACGGCCAUUUCCACCGGCCAACUUAUGGGCCACAUGG ........-----........((((((.....----------------(((((((((((.........)))))))).........((((.........))))..)))...)))))).... ( -29.70) >DroYak_CAF1 12566 120 - 1 UCAACCCCCCUGUUUUUCUUCGUGGCCAUUCAACAUUCAACAUUGAAAAGGGGAUUGGGCGACACGUACUCGGUCCAUACAUUACGGCCAUUUCCACCGGCCAACUUAUGGGCCACAUGG .......((.(((........((.(((..((..(.((((....))))..)..))...))).))........((((((((......((((.........))))....))))))))))).)) ( -31.70) >consensus UCAACCCC_____UUUUCUUCGUGGCCAUUCA_______________AAAGGGAUUGGGCGACACGUACUCCGUCCAUACAUUACGGCCAUUUCCACCGGCCAACUUAUGGGCCACAUGG .....................((((((........................((((.(((.........))).)))).........((((.........))))........)))))).... (-25.36 = -25.56 + 0.20)

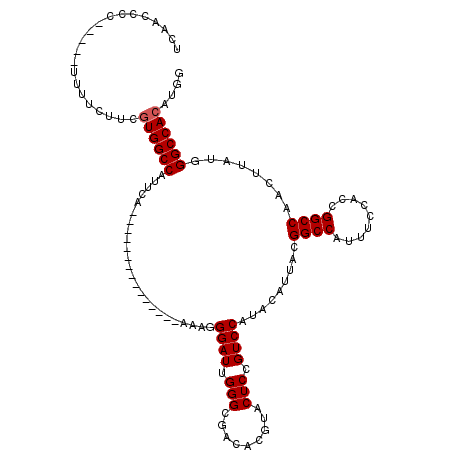

| Location | 4,654,767 – 4,654,874 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.80 |
| Mean single sequence MFE | -32.48 |
| Consensus MFE | -30.68 |
| Energy contribution | -31.48 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.94 |
| SVM decision value | 4.15 |
| SVM RNA-class probability | 0.999817 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4654767 107 + 22224390 --------UGAAUGGCCACGAAGAAAA-----GGGGUUGAGACGAAAGACUUUCGAAGGAAGGGACUUUAAUUAUGAGCCCGACUCGGGCUAUUAAAACAACUCUCUGGCCAUAUCUAAU --------.(((((((((.((......-----.((((((.....((((.(((((....)))))..)))).......((((((...)))))).......))))))))))))))).)).... ( -34.81) >DroSec_CAF1 16447 107 + 1 --------UGAAUGGCCAUGAAGAAAA-----GGGGUUGAGACGAAAGACUUGCGAAGGAAGGGACUUUAAUUAUGAGCCCGACCCGGGCUAUUAAAACAACUCUCUGGCCAUAUCUAAU --------.(((((((((.((......-----.((((((.....((((.(((.(....).)))..)))).......((((((...)))))).......))))))))))))))).)).... ( -30.41) >DroSim_CAF1 5103 107 + 1 --------UGAAUGGCCACGAAGAAAA-----GGGGUUGAGACGAAAGACUUGCGAAGGAAGGGACUUUAAUUAUGAGCCCGACCCGGGCUAUUAAAACAACUCUCUGGCCAUAUCUAAU --------.(((((((((.((......-----.((((((.....((((.(((.(....).)))..)))).......((((((...)))))).......))))))))))))))).)).... ( -30.41) >DroEre_CAF1 17026 107 + 1 --------UGAAUGGCCACGAAGAAAA-----GGGGUUGAGGCGAAAGACUUUCCAAGGAAGGGACUUUAAUUAUGAGCCCGACCCGGGCUAUUAAAACAACUCUCCGGCCAUAUCUAAU --------.((((((((..........-----.((((((.(((.((((.(((((....)))))..))))........)))))))))(((...............))))))))).)).... ( -33.06) >DroYak_CAF1 12646 120 + 1 UUGAAUGUUGAAUGGCCACGAAGAAAAACAGGGGGGUUGAGACGCAAGACUUUCCAAAGAAGGGACUUUAAUUAUGAGCCCGACCCGGGCUAUUAAAACAACUCUUUGGCCAUAUCUAAU .........((((((((.............((..((((.........))))..))...((((((..((((((....((((((...))))))))))))....)))))))))))).)).... ( -33.70) >consensus ________UGAAUGGCCACGAAGAAAA_____GGGGUUGAGACGAAAGACUUUCGAAGGAAGGGACUUUAAUUAUGAGCCCGACCCGGGCUAUUAAAACAACUCUCUGGCCAUAUCUAAU .........(((((((((..............(((((((.....((((.(((((....)))))..)))).......((((((...)))))).......))))))).))))))).)).... (-30.68 = -31.48 + 0.80)


| Location | 4,654,767 – 4,654,874 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.80 |
| Mean single sequence MFE | -27.24 |
| Consensus MFE | -23.68 |
| Energy contribution | -23.36 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.814632 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4654767 107 - 22224390 AUUAGAUAUGGCCAGAGAGUUGUUUUAAUAGCCCGAGUCGGGCUCAUAAUUAAAGUCCCUUCCUUCGAAAGUCUUUCGUCUCAACCCC-----UUUUCUUCGUGGCCAUUCA-------- ....((.((((((((((((..((((((((((((((...))))))....)))))))..........((((.....)))).........)-----..)))))..))))))))).-------- ( -26.80) >DroSec_CAF1 16447 107 - 1 AUUAGAUAUGGCCAGAGAGUUGUUUUAAUAGCCCGGGUCGGGCUCAUAAUUAAAGUCCCUUCCUUCGCAAGUCUUUCGUCUCAACCCC-----UUUUCUUCAUGGCCAUUCA-------- ....((.((((((((((((..((((((((((((((...))))))....)))))))..........(....)................)-----..)))))..))))))))).-------- ( -26.00) >DroSim_CAF1 5103 107 - 1 AUUAGAUAUGGCCAGAGAGUUGUUUUAAUAGCCCGGGUCGGGCUCAUAAUUAAAGUCCCUUCCUUCGCAAGUCUUUCGUCUCAACCCC-----UUUUCUUCGUGGCCAUUCA-------- ....((.((((((((((((..((((((((((((((...))))))....)))))))..........(....)................)-----..)))))..))))))))).-------- ( -26.00) >DroEre_CAF1 17026 107 - 1 AUUAGAUAUGGCCGGAGAGUUGUUUUAAUAGCCCGGGUCGGGCUCAUAAUUAAAGUCCCUUCCUUGGAAAGUCUUUCGCCUCAACCCC-----UUUUCUUCGUGGCCAUUCA-------- ....((.(((((((((.((..((((((((((((((...))))))....))))))))..)))))..((((((................)-----))))).....)))))))).-------- ( -27.99) >DroYak_CAF1 12646 120 - 1 AUUAGAUAUGGCCAAAGAGUUGUUUUAAUAGCCCGGGUCGGGCUCAUAAUUAAAGUCCCUUCUUUGGAAAGUCUUGCGUCUCAACCCCCCUGUUUUUCUUCGUGGCCAUUCAACAUUCAA ....((.(((((((..(((.......(((((...((((.((((.((..(((....(((.......))).)))..)).))).).))))..)))))...)))..)))))))))......... ( -29.40) >consensus AUUAGAUAUGGCCAGAGAGUUGUUUUAAUAGCCCGGGUCGGGCUCAUAAUUAAAGUCCCUUCCUUCGAAAGUCUUUCGUCUCAACCCC_____UUUUCUUCGUGGCCAUUCA________ ....((.((((((((((((...(((((((((((((...))))))....)))))))...((((....).)))........................)))))..)))))))))......... (-23.68 = -23.36 + -0.32)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:15:21 2006