
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,650,431 – 4,650,641 |
| Length | 210 |
| Max. P | 0.983795 |

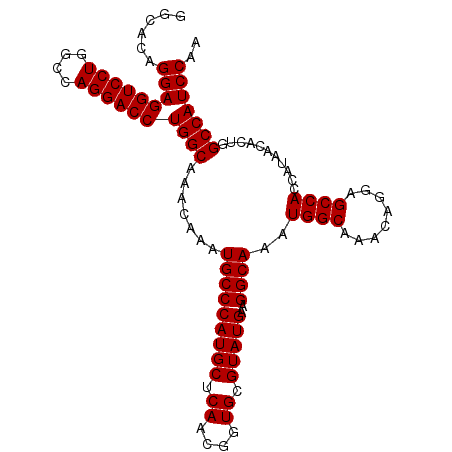
| Location | 4,650,431 – 4,650,533 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 96.15 |
| Mean single sequence MFE | -36.83 |
| Consensus MFE | -35.10 |
| Energy contribution | -35.10 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.98 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.95 |
| SVM RNA-class probability | 0.983795 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4650431 102 - 22224390 --UACUGGAGGUCCUGGCCAGGACCUGGCAAACAAAUGCCCAUGCUCAACGGUGCGUAUGAAGGCAAAUGGCAAACAGGAGCCACCAUAACACUAGCCAUCCAA --...((((((((((....))))))((((.......(((((((((.((....)).)))))..))))..((((........))))...........)))))))). ( -35.90) >DroEre_CAF1 12705 104 - 1 GGCACAGGAGGUCCUGGCCAGGACCUGGCAAACAAAUGCCCAUGCUCAACGGUGCGUAUGAAGGCAAAUGGCAAACAGGAGCCACCAUAACACUGGCCAUCCGA (((.(((.(((((((....)))))))((........(((((((((.((....)).)))))..))))..((((........))))))......))))))...... ( -37.30) >DroYak_CAF1 8129 104 - 1 GGCACAGGAGGUCCUGGCCAGGACCUGGCAAACAAAUGCCCAUGCUCAACGGUGCGUAUGAAGGCAAAUGGCAAACAGGAGCCACCAUAACACUGGCCAUCCAA (((.(((.(((((((....)))))))((........(((((((((.((....)).)))))..))))..((((........))))))......))))))...... ( -37.30) >consensus GGCACAGGAGGUCCUGGCCAGGACCUGGCAAACAAAUGCCCAUGCUCAACGGUGCGUAUGAAGGCAAAUGGCAAACAGGAGCCACCAUAACACUGGCCAUCCAA ......(((((((((....))))))((((.......(((((((((.((....)).)))))..))))..((((........))))...........))))))).. (-35.10 = -35.10 + 0.00)


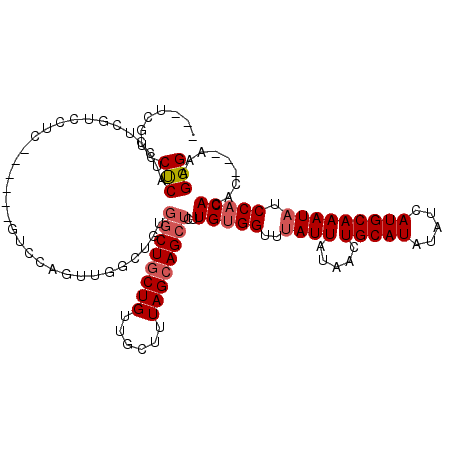
| Location | 4,650,533 – 4,650,641 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.36 |
| Mean single sequence MFE | -29.12 |
| Consensus MFE | -18.08 |
| Energy contribution | -18.92 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.650642 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4650533 108 + 22224390 ---UCGUUCUCAUCCUCAACCUC------GUCCAGUUGGCUGUGGCUGCUGUUGCUUUAGCAGCUCUUGUGGUUUAUUAUAACUGCAUAUAUCAUGCAAAUAUCCACACAC---AAAGGG ---..........(((((((...------.....))))..(((((((((((......)))))))...(((((..((((.....(((((.....))))))))).))))))))---).))). ( -28.70) >DroSim_CAF1 56 111 + 1 UCCUCAUCCUCAUCCUCGUCCUC------GUCCAGUUGGCUGUGGCUGCUGUUGCUUUAGCAGCUCUUGUGGUUUAUUAUAACUGCAUAUAUCAUGCAAAUAUCCACACAC---AAAGGG .............(((.(((..(------.....)..)))(((((((((((......)))))))...(((((..((((.....(((((.....))))))))).))))))))---).))). ( -28.60) >DroEre_CAF1 12809 114 + 1 ---UCGCCCUCGUCCUCGUCCUCGUAGUCGUCCAUUUGGCUGUGGCUGCUGUUGCUUUAGCAGCUCUUGUGGUUUAUUAUAACUGCAUAUAUCAUGCAAAUAUCCACACAC---AGAGGG ---...(((((..............(((((......)))))((((((((((......)))))))...(((((..((((.....(((((.....))))))))).))))))))---.))))) ( -36.00) >DroYak_CAF1 8233 108 + 1 ---UCGUCCUCGUCCCCGUCCCC------GUCUAGUUGGCUGUGGCUGCUGUUGCUUUAGCAGCUCUUGUGGUUUAUUAUAACUGCAUAUAUCAUGCAAAUCUCCACACAC---AGAGGG ---....((((............------(((.....)))(((((((((((......)))))))...(((((...(((.....(((((.....))))))))..))))))))---))))). ( -31.40) >DroAna_CAF1 15731 98 + 1 --------CCCACCUUCGACCU-------------UCGACCGC-CCUUCUGUUGCUUUAGCAGCUCUUGUGGUUUAUUAUACCUGCAUAUAUCAUGCAAAUAUCCUCACAGCUGACAGGG --------.......(((....-------------.)))...(-(((((.(((((....)))))..((((((..((((.....(((((.....)))))))))..).)))))..)).)))) ( -20.90) >consensus ___UCGUCCUCAUCCUCGUCCUC______GUCCAGUUGGCUGUGGCUGCUGUUGCUUUAGCAGCUCUUGUGGUUUAUUAUAACUGCAUAUAUCAUGCAAAUAUCCACACAC___AAAGGG ........(((................................((((((((......))))))))..(((((..((((.....(((((.....))))))))).))))).........))) (-18.08 = -18.92 + 0.84)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:15:12 2006