
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,638,282 – 4,638,420 |
| Length | 138 |
| Max. P | 0.997347 |

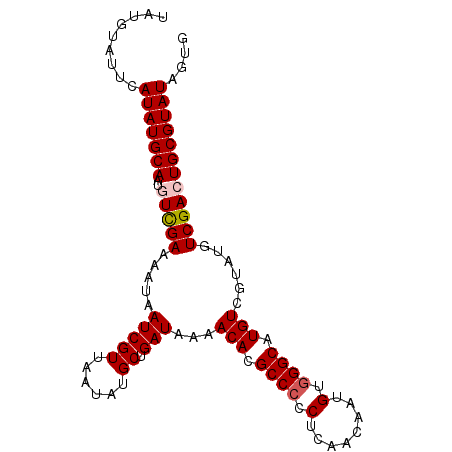
| Location | 4,638,282 – 4,638,380 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 96.33 |
| Mean single sequence MFE | -23.74 |
| Consensus MFE | -19.98 |
| Energy contribution | -20.42 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.638876 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4638282 98 - 22224390 UAUGUAUUCAUAUGCAAUGUCGAAAAUAAUCGUUAAUAUGCUGAUAAAACACGCCCCCUCAACAUUGUGGGCAUGUCGUUUGUCGACUGCGUAUAGUG .........(((((((..(((((((((.(((((......)).)))...(((.((((.(........).)))).))).)))).)))))))))))).... ( -23.00) >DroSec_CAF1 23992 98 - 1 UAUGUAUUCAUAUGCAAUGUCGAAAAUAAUCGUUAAUAUGCUGAUAAAACACGCCCCCUCAACAAUGUGGGCAUGUCGUGUGUCGACUGCGUAUAGUG .........(((((((..(((((.....(((((......)).)))...((((((.(((.((....)).)))...).))))).)))))))))))).... ( -23.70) >DroSim_CAF1 5623 98 - 1 UAUGUAUUCAUAUGCAAUGUCGAAAAUAAUCGUUAAUAUGCUGAUAAAACACGCCCCCUCAACAAUGUGGGCAUGUCGUGUGUCGACUGCGUAUAGUG .........(((((((..(((((.....(((((......)).)))...((((((.(((.((....)).)))...).))))).)))))))))))).... ( -23.70) >DroEre_CAF1 22082 98 - 1 UAUGUAUUCAUAUGCAAUGUCGAAAAUAAUCGUUAAUAUGCUGAUAAAACACGCCCCCUCAACAAUGUGGGCAUGUCGAAUGUCGAAUGCGUAUAGUG (((((((((....(((((..(((......)))...)).))).((((..(((.((((.(........).)))).)))....)))))))))))))..... ( -24.10) >DroYak_CAF1 23459 97 - 1 UAUGUAUUCAUAUGCAAUGUUGAAAAUAAUCGUUAAUAUGCUGAUAAAACACGCC-CCUCAACAAUGUGGGCAUGUCGAAUGUCGAAUGCGUAUAGUA (((((((((....(((.((((((.........))))))))).((((..(((.(((-((........).)))).)))....)))))))))))))..... ( -24.20) >consensus UAUGUAUUCAUAUGCAAUGUCGAAAAUAAUCGUUAAUAUGCUGAUAAAACACGCCCCCUCAACAAUGUGGGCAUGUCGUAUGUCGACUGCGUAUAGUG .........(((((((..(((((.....(((((......)).)))...(((.((((.(........).)))).)))......)))))))))))).... (-19.98 = -20.42 + 0.44)

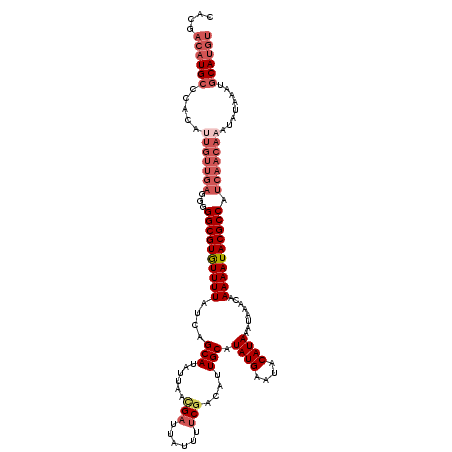
| Location | 4,638,300 – 4,638,420 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.67 |
| Mean single sequence MFE | -29.56 |
| Consensus MFE | -24.83 |
| Energy contribution | -26.92 |
| Covariance contribution | 2.09 |
| Combinations/Pair | 1.06 |
| Mean z-score | -4.11 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.84 |
| SVM RNA-class probability | 0.997347 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
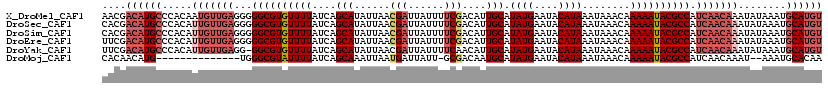
>X_DroMel_CAF1 4638300 120 + 22224390 AACGACAUGCCCACAAUGUUGAGGGGGCGUGUUUUAUCAGCAUAUUAACGAUUAUUUUCGACAUUGCAUAUGAAUACAUAAAUAAACAAAAAUACGCCAUCAACAAAUAUAAAUGCAUGU ....((((((......((((((...((((((((((....(((......(((......)))....))).((((....))))........)))))))))).)))))).........)))))) ( -31.46) >DroSec_CAF1 24010 120 + 1 CACGACAUGCCCACAUUGUUGAGGGGGCGUGUUUUAUCAGCAUAUUAACGAUUAUUUUCGACAUUGCAUAUGAAUACAUAAAUAAACAAAAAUACGCCAUCAACAAAUAUAAAUGCAUGU ....((((((.....(((((((...((((((((((....(((......(((......)))....))).((((....))))........)))))))))).)))))))........)))))) ( -32.52) >DroSim_CAF1 5641 120 + 1 CACGACAUGCCCACAUUGUUGAGGGGGCGUGUUUUAUCAGCAUAUUAACGAUUAUUUUCGACAUUGCAUAUGAAUACAUAAAUAAACAAAAAUACGCCAUCAACAAAUAUAAAUGCAUGU ....((((((.....(((((((...((((((((((....(((......(((......)))....))).((((....))))........)))))))))).)))))))........)))))) ( -32.52) >DroEre_CAF1 22100 120 + 1 UUCGACAUGCCCACAUUGUUGAGGGGGCGUGUUUUAUCAGCAUAUUAACGAUUAUUUUCGACAUUGCAUAUGAAUACAUAAAUAAACAAAAAUACGCCAUCAACAAAUAUAAAUGCAUGU ....((((((.....(((((((...((((((((((....(((......(((......)))....))).((((....))))........)))))))))).)))))))........)))))) ( -32.52) >DroYak_CAF1 23477 119 + 1 UUCGACAUGCCCACAUUGUUGAGG-GGCGUGUUUUAUCAGCAUAUUAACGAUUAUUUUCAACAUUGCAUAUGAAUACAUAAAUAAACAAAAAUACGCCAUCAACAAAUAUAAAUGCAUGU ....((((((.....(((((((..-((((((((((....(((.......((......)).....))).((((....))))........)))))))))).)))))))........)))))) ( -29.92) >DroMoj_CAF1 116009 103 + 1 CACAACAUG--------------UGGGCGUAUUUUAUCAGCAAAUUAAUGAUUAUU-GCGACAAUGCAUAUGAAUACAUAAAUAAACAAAAAUACGCCAUCAACAAAU--AAAUGCACAA (((.....)--------------))((((((((((....((((...........))-)).........((((....))))........))))))))))..........--.......... ( -18.40) >consensus CACGACAUGCCCACAUUGUUGAGGGGGCGUGUUUUAUCAGCAUAUUAACGAUUAUUUUCGACAUUGCAUAUGAAUACAUAAAUAAACAAAAAUACGCCAUCAACAAAUAUAAAUGCAUGU ....((((((.....(((((((...((((((((((....(((......(((......)))....))).((((....))))........)))))))))).)))))))........)))))) (-24.83 = -26.92 + 2.09)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:15:05 2006