
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,636,931 – 4,637,122 |
| Length | 191 |
| Max. P | 0.747318 |

| Location | 4,636,931 – 4,637,045 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.22 |
| Mean single sequence MFE | -19.82 |
| Consensus MFE | -16.12 |
| Energy contribution | -16.12 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.588520 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4636931 114 + 22224390 AAUCUGGCAACA------GCAACAAAGUGAAUAAAGUGAUGACAAAAGUAAACUGUAUCCAAAACAAAACCCCAAUGAAGUUUAUGAUUUCGAUUCUGCUCAACUAGAGAAUCGAAUUCA ...(((....))------)............................(((((((........................)))))))((.(((((((((.((.....))))))))))).)). ( -19.86) >DroSec_CAF1 23067 114 + 1 AAUCUGGCAACA------GCAACAAAGUGAAUAAAGUGAUGACAAAAGUAAACUGUAUCCAAAACAAAACCCCAAUGAAGUUUAUGAUUUCGAUUCUGCUCAACUAGAGAAUCGAAUUCA ...(((....))------)............................(((((((........................)))))))((.(((((((((.((.....))))))))))).)). ( -19.86) >DroSim_CAF1 4686 114 + 1 AAUCUGGCAACA------GCAACAAAGUGAAUAAAGUGAUGACAAAAGUAAACUGUAUCCAAAACAAAACCCCAAUGAAGUUUAUGAUUUCGAUUCUGCUCAACUAGAGAAUCGAAUUCA ...(((....))------)............................(((((((........................)))))))((.(((((((((.((.....))))))))))).)). ( -19.86) >DroEre_CAF1 21135 114 + 1 AAUCUGGCAACA------GCAACAAAGUGAAUAAAGUGAUGACAAAAGUAAACUGUAUCCAAAACAAAACCCCAAUGAAGUUUAUGAUUUCGAUUCUGUUCAACUAGAGAAUCGAAUUCA ...(((....))------)............................(((((((........................)))))))((.(((((((((.(......).))))))))).)). ( -20.56) >DroYak_CAF1 22459 120 + 1 AAUCUGGCAACAGCAACAGCAACAAAGUGAAUAAAGUGAUGACAAAAGUAAACUGUAUCCAAAACAAACCCAUAAUGAAGUUUAUGAUUUCGAUUCUGCCCAACUAGAGAAUCGAAUUCA ...(((....)))..................................(((((((........................)))))))((.(((((((((..........))))))))).)). ( -18.96) >consensus AAUCUGGCAACA______GCAACAAAGUGAAUAAAGUGAUGACAAAAGUAAACUGUAUCCAAAACAAAACCCCAAUGAAGUUUAUGAUUUCGAUUCUGCUCAACUAGAGAAUCGAAUUCA ....((....))...................................(((((((........................)))))))((.(((((((((..........))))))))).)). (-16.12 = -16.12 + -0.00)

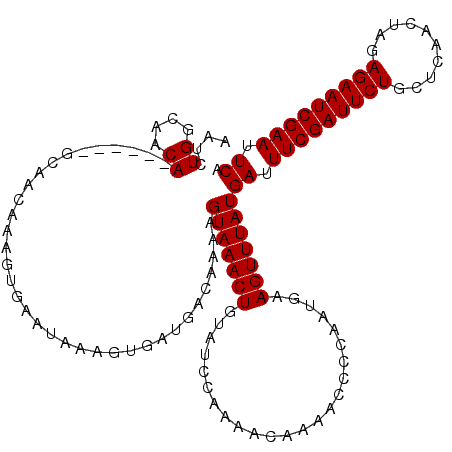
| Location | 4,637,005 – 4,637,122 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.49 |
| Mean single sequence MFE | -23.36 |
| Consensus MFE | -17.60 |
| Energy contribution | -18.08 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.747318 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4637005 117 + 22224390 UUUAUGAUUUCGAUUCUGCUCAACUAGAGAAUCGAAUUCAACUGUAGGUAUAUUAACAUUGAAUGCAAUUC-GUGCAGAGG--GCACACAACUAAAUUGUACAAUACAAAAAAUAUAUAA ....(((.(((((((((.((.....))))))))))).)))..((((.(((((........((((...))))-((((.....--))))..........)))))..))))............ ( -24.10) >DroSec_CAF1 23141 115 + 1 UUUAUGAUUUCGAUUCUGCUCAACUAGAGAAUCGAAUUCAACUGUAGGUAUAUUAGCAUUGAAUGCAAUUC-GUGCAGAGGGAGCACACAACUAAACUGUACAAUAAAA----AAUAUAA ....(((.(((((((((.((.....))))))))))).)))........((((((.((((...)))).....-((((((....((.......))...)))))).......----)))))). ( -24.90) >DroSim_CAF1 4760 115 + 1 UUUAUGAUUUCGAUUCUGCUCAACUAGAGAAUCGAAUUCAACUGUAGGUAUAUUAACAUUGAAUGCGAUUC-GUGCAGAGGGAGCACACAACUAAACUGUACAAUAAAA----AAUAUAA ....(((.(((((((((.((.....))))))))))).))).......(((((......(((..(((..(((-.....)))...)))..)))......))))).......----....... ( -22.50) >DroEre_CAF1 21209 114 + 1 UUUAUGAUUUCGAUUCUGUUCAACUAGAGAAUCGAAUUCAACUGUAGGUAUAUUAACAUUGAAUGCAAUUCAAUGCAGAGG--GCACGAAACUAAAUUGUACAAUAAAA----UAAAAAA ....(((.(((((((((.(......).))))))))).))).......(((((....((((((((...))))))))....((--........))....))))).......----....... ( -22.30) >DroYak_CAF1 22539 113 + 1 UUUAUGAUUUCGAUUCUGCCCAACUAGAGAAUCGAAUUCAACUGUAGGUAUAUUAACAUUGCAUGCCAUUC-AUGCUGAGG--GCACAAAACUAAACUGUACAAUAAAA----UAUAUAA ....(((.(((((((((..........))))))))).))).......(((((((...((((..((((.(((-.....))))--)))....((......)).))))..))----))))).. ( -23.00) >consensus UUUAUGAUUUCGAUUCUGCUCAACUAGAGAAUCGAAUUCAACUGUAGGUAUAUUAACAUUGAAUGCAAUUC_GUGCAGAGG__GCACACAACUAAACUGUACAAUAAAA____UAUAUAA ....(((.(((((((((..........))))))))).))).......(((((.....((((....))))...((((.......))))..........))))).................. (-17.60 = -18.08 + 0.48)

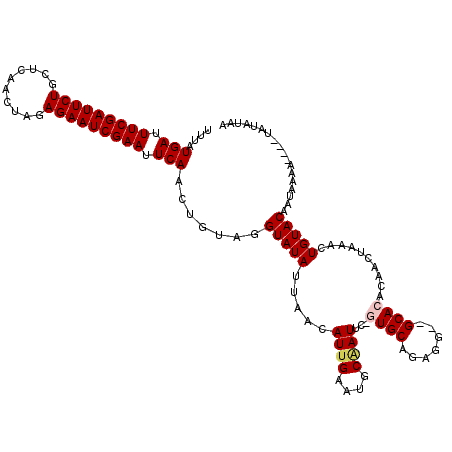

| Location | 4,637,005 – 4,637,122 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.49 |
| Mean single sequence MFE | -23.60 |
| Consensus MFE | -18.86 |
| Energy contribution | -19.06 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.732014 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4637005 117 - 22224390 UUAUAUAUUUUUUGUAUUGUACAAUUUAGUUGUGUGC--CCUCUGCAC-GAAUUGCAUUCAAUGUUAAUAUACCUACAGUUGAAUUCGAUUCUCUAGUUGAGCAGAAUCGAAAUCAUAAA ...((((((..(((...((((((((...)))))((((--.....))))-.....)))..)))....))))))........(((.(((((((((((.....)).))))))))).))).... ( -23.60) >DroSec_CAF1 23141 115 - 1 UUAUAUU----UUUUAUUGUACAGUUUAGUUGUGUGCUCCCUCUGCAC-GAAUUGCAUUCAAUGCUAAUAUACCUACAGUUGAAUUCGAUUCUCUAGUUGAGCAGAAUCGAAAUCAUAAA .((((((----...(((((...(((.(((((.(((((.......))))-)))))).))))))))..))))))........(((.(((((((((((.....)).))))))))).))).... ( -24.60) >DroSim_CAF1 4760 115 - 1 UUAUAUU----UUUUAUUGUACAGUUUAGUUGUGUGCUCCCUCUGCAC-GAAUCGCAUUCAAUGUUAAUAUACCUACAGUUGAAUUCGAUUCUCUAGUUGAGCAGAAUCGAAAUCAUAAA .((((((----...(((((.((((.....))))((((.......))))-..........)))))..))))))........(((.(((((((((((.....)).))))))))).))).... ( -23.80) >DroEre_CAF1 21209 114 - 1 UUUUUUA----UUUUAUUGUACAAUUUAGUUUCGUGC--CCUCUGCAUUGAAUUGCAUUCAAUGUUAAUAUACCUACAGUUGAAUUCGAUUCUCUAGUUGAACAGAAUCGAAAUCAUAAA .......----....((((((.......((.....))--.....(((((((((...))))))))).........))))))(((.(((((((((..........))))))))).))).... ( -19.60) >DroYak_CAF1 22539 113 - 1 UUAUAUA----UUUUAUUGUACAGUUUAGUUUUGUGC--CCUCAGCAU-GAAUGGCAUGCAAUGUUAAUAUACCUACAGUUGAAUUCGAUUCUCUAGUUGGGCAGAAUCGAAAUCAUAAA ...((((----((.(((((((..(((....((..(((--.....))).-.)).))).)))))))..))))))........(((.(((((((((((.....)).))))))))).))).... ( -26.40) >consensus UUAUAUA____UUUUAUUGUACAGUUUAGUUGUGUGC__CCUCUGCAC_GAAUUGCAUUCAAUGUUAAUAUACCUACAGUUGAAUUCGAUUCUCUAGUUGAGCAGAAUCGAAAUCAUAAA ...............((((((..(((..((((.((((....((......))...)))).))))...))).....))))))(((.(((((((((((.....)).))))))))).))).... (-18.86 = -19.06 + 0.20)

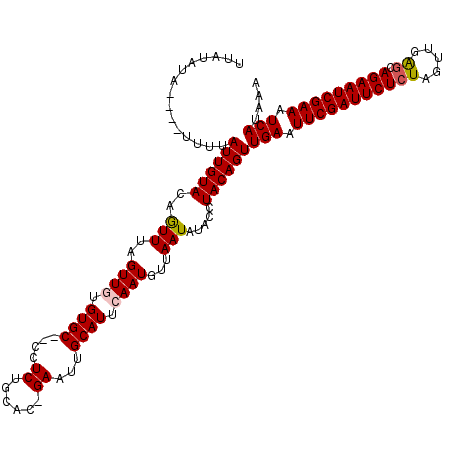
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:15:03 2006