
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,630,937 – 4,631,161 |
| Length | 224 |
| Max. P | 0.981341 |

| Location | 4,630,937 – 4,631,043 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.44 |
| Mean single sequence MFE | -26.15 |
| Consensus MFE | -18.89 |
| Energy contribution | -20.77 |
| Covariance contribution | 1.88 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.780398 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4630937 106 + 22224390 ACUAACGCACAGACAAUCUGCUGCGGAAUGGCCGCAGAUAUAGAAA------CUGAAACUAAAACCGACGAACGGCGGCCAUCAGGAC--------AUCAAUCAAAUGGCCGCAUGUCCU .(((.....(((.....)))((((((.....))))))...)))...------..............((((....((((((((...((.--------.....))..)))))))).)))).. ( -31.30) >DroSec_CAF1 17361 106 + 1 ACUAAC--ACAGACAAUCUGUUGCGGAAUGGCCGCCGACAUAGCAACUGA-ACUGAAACUAAAACC---GAACGGCGGCCAUCAGGAC--------AUCAAUCAAAUGGCCGCAUGUCCU ......--...((((.((.(((((...(((........))).))))).))-...............---.....((((((((...((.--------.....))..)))))))).)))).. ( -28.50) >DroEre_CAF1 15322 107 + 1 ACCAGA--ACAGACAAUCUGCUGCAGAAUAGCCACAGACAUAGAAACUGAAACUGAAACUAAAACC---GAACGGCGGCCAUCGGGAC--------AUCAAUCAAAUGGCCGCAUGUCCU ......--...((((.((((..((......))..))))............................---.....((((((((...((.--------.....))..)))))))).)))).. ( -24.80) >DroYak_CAF1 15815 106 + 1 ACUA-A--ACACACAGUCUGCUG--------CCACAGACAUAGAAGCUGAAACUGAAACUAAAACC---GAACGACGGCCAUCGGGACAUCAGGACAUCAAUCAAAUGGCCCCAUGUCCU ....-.--.......(((((...--------...))))).........................((---(.....))).....(((((((..((.(((.......))).))..))))))) ( -20.00) >consensus ACUAAA__ACAGACAAUCUGCUGCGGAAUGGCCACAGACAUAGAAACUGA_ACUGAAACUAAAACC___GAACGGCGGCCAUCAGGAC________AUCAAUCAAAUGGCCGCAUGUCCU ...........((((((((((.((......)).))))))...................................((((((((...((..............))..)))))))).)))).. (-18.89 = -20.77 + 1.88)


| Location | 4,630,937 – 4,631,043 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.44 |
| Mean single sequence MFE | -34.70 |
| Consensus MFE | -24.12 |
| Energy contribution | -24.50 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.775856 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4630937 106 - 22224390 AGGACAUGCGGCCAUUUGAUUGAU--------GUCCUGAUGGCCGCCGUUCGUCGGUUUUAGUUUCAG------UUUCUAUAUCUGCGGCCAUUCCGCAGCAGAUUGUCUGUGCGUUAGU .((((..(((((((((.(((....--------)))..))))))))).))))..........((..(((------..(((....((((((.....)))))).)))....))).))...... ( -34.90) >DroSec_CAF1 17361 106 - 1 AGGACAUGCGGCCAUUUGAUUGAU--------GUCCUGAUGGCCGCCGUUC---GGUUUUAGUUUCAGU-UCAGUUGCUAUGUCGGCGGCCAUUCCGCAACAGAUUGUCUGU--GUUAGU ..(((.(((((.((((.....)))--------)....(((((((((((..(---(....((((..(...-...)..)))))).))))))))))))))))((((.....))))--)))... ( -36.00) >DroEre_CAF1 15322 107 - 1 AGGACAUGCGGCCAUUUGAUUGAU--------GUCCCGAUGGCCGCCGUUC---GGUUUUAGUUUCAGUUUCAGUUUCUAUGUCUGUGGCUAUUCUGCAGCAGAUUGUCUGU--UCUGGU .((((..(((((((((.(((....--------)))..))))))))).))))---..........((((...(((...(...((((((.((......)).)))))).).))).--.)))). ( -36.40) >DroYak_CAF1 15815 106 - 1 AGGACAUGGGGCCAUUUGAUUGAUGUCCUGAUGUCCCGAUGGCCGUCGUUC---GGUUUUAGUUUCAGUUUCAGCUUCUAUGUCUGUGG--------CAGCAGACUGUGUGU--U-UAGU .((((((.(((.((((.....)))).))).))))))(...(((((.....)---))))...)...((((((..(((.((((....))))--------.))))))))).....--.-.... ( -31.50) >consensus AGGACAUGCGGCCAUUUGAUUGAU________GUCCCGAUGGCCGCCGUUC___GGUUUUAGUUUCAGU_UCAGUUUCUAUGUCUGCGGCCAUUCCGCAGCAGAUUGUCUGU__GUUAGU .(((((.(((((((((.(((............)))..)))))))))...................................((((((.((......)).))))))))))).......... (-24.12 = -24.50 + 0.38)


| Location | 4,630,977 – 4,631,083 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.65 |
| Mean single sequence MFE | -32.53 |
| Consensus MFE | -26.47 |
| Energy contribution | -27.21 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.815256 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4630977 106 + 22224390 UAGAAA------CUGAAACUAAAACCGACGAACGGCGGCCAUCAGGAC--------AUCAAUCAAAUGGCCGCAUGUCCUUUGGCCUGGUCGGGCCAUUUAAUUUCGCAUUUGUGCCAUU ..((((------.((((.........((((....((((((((...((.--------.....))..)))))))).))))...((((((....)))))))))).))))(((....))).... ( -33.70) >DroSec_CAF1 17399 108 + 1 UAGCAACUGA-ACUGAAACUAAAACC---GAACGGCGGCCAUCAGGAC--------AUCAAUCAAAUGGCCGCAUGUCCUUUGGCCUGGUCGGGCCAUUUAAUUUCGCAUUUGUGCCAUU ..(((.....-..(((((.((((...---((...((((((((...((.--------.....))..))))))))...))...((((((....)))))))))).)))))......))).... ( -33.22) >DroEre_CAF1 15360 108 + 1 UAGAAACUGAAACUGAAACUAAAACC---GAACGGCGGCCAUCGGGAC--------AUCAAUCAAAUGGCCGCAUGUCCUUUGGCCUGGUCG-GCCAUUUAAUUUCGCAUUUGUGCCAUU ..((((.((((...............---((...((((((((...((.--------.....))..))))))))...))...(((((.....)-)))))))).))))(((....))).... ( -30.60) >DroYak_CAF1 15844 117 + 1 UAGAAGCUGAAACUGAAACUAAAACC---GAACGACGGCCAUCGGGACAUCAGGACAUCAAUCAAAUGGCCCCAUGUCCUUUGGCCUGGUCGGGCCAUUUAAUUUCGCAUUUGUGCCAUU ..............((((.((((...---(..(((((((((..(((((((..((.(((.......))).))..))))))).)))))..))))..)..)))).))))(((....))).... ( -32.60) >consensus UAGAAACUGA_ACUGAAACUAAAACC___GAACGGCGGCCAUCAGGAC________AUCAAUCAAAUGGCCGCAUGUCCUUUGGCCUGGUCGGGCCAUUUAAUUUCGCAUUUGUGCCAUU ..............((((.((((......((...((((((((...((..............))..))))))))...))...((((((....)))))))))).))))(((....))).... (-26.47 = -27.21 + 0.75)


| Location | 4,630,977 – 4,631,083 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.65 |
| Mean single sequence MFE | -40.42 |
| Consensus MFE | -32.44 |
| Energy contribution | -32.75 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.25 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.88 |
| SVM RNA-class probability | 0.981341 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4630977 106 - 22224390 AAUGGCACAAAUGCGAAAUUAAAUGGCCCGACCAGGCCAAAGGACAUGCGGCCAUUUGAUUGAU--------GUCCUGAUGGCCGCCGUUCGUCGGUUUUAGUUUCAG------UUUCUA ....(((....)))((((((((((((((......)))))..((((..(((((((((.(((....--------)))..))))))))).))))......)))))))))..------...... ( -38.90) >DroSec_CAF1 17399 108 - 1 AAUGGCACAAAUGCGAAAUUAAAUGGCCCGACCAGGCCAAAGGACAUGCGGCCAUUUGAUUGAU--------GUCCUGAUGGCCGCCGUUC---GGUUUUAGUUUCAGU-UCAGUUGCUA ..(((((.....((((((((((((((((......)))))..((((..(((((((((.(((....--------)))..))))))))).))))---...))))))))).))-.....))))) ( -41.50) >DroEre_CAF1 15360 108 - 1 AAUGGCACAAAUGCGAAAUUAAAUGGC-CGACCAGGCCAAAGGACAUGCGGCCAUUUGAUUGAU--------GUCCCGAUGGCCGCCGUUC---GGUUUUAGUUUCAGUUUCAGUUUCUA ....(((....)))(((((((((((((-(.....)))))..((((..(((((((((.(((....--------)))..))))))))).))))---...))))))))).............. ( -39.00) >DroYak_CAF1 15844 117 - 1 AAUGGCACAAAUGCGAAAUUAAAUGGCCCGACCAGGCCAAAGGACAUGGGGCCAUUUGAUUGAUGUCCUGAUGUCCCGAUGGCCGUCGUUC---GGUUUUAGUUUCAGUUUCAGCUUCUA ...(((..((((..(((((((((..(((.(((..(((((..((((((.(((.((((.....)))).))).))))))...)))))...))).---)))))))))))).))))..))).... ( -42.30) >consensus AAUGGCACAAAUGCGAAAUUAAAUGGCCCGACCAGGCCAAAGGACAUGCGGCCAUUUGAUUGAU________GUCCCGAUGGCCGCCGUUC___GGUUUUAGUUUCAGU_UCAGUUUCUA ....(((....)))((((((((((((((......)))))..((((..(((((((((.(((............)))..))))))))).))))......))))))))).............. (-32.44 = -32.75 + 0.31)


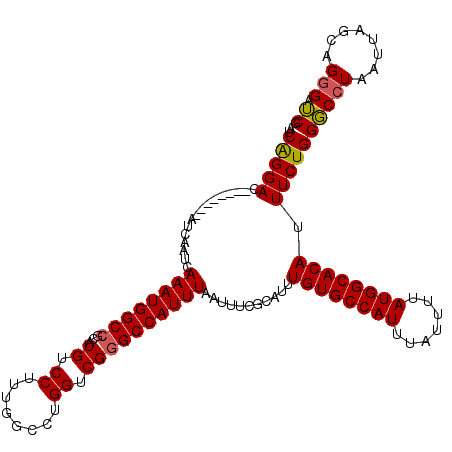
| Location | 4,631,011 – 4,631,123 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.39 |
| Mean single sequence MFE | -35.12 |
| Consensus MFE | -28.77 |
| Energy contribution | -28.65 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.831506 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4631011 112 + 22224390 AUCAGGAC--------AUCAAUCAAAUGGCCGCAUGUCCUUUGGCCUGGUCGGGCCAUUUAAUUUCGCAUUUGUGCCAUUUAUUUUAUGGCACAUUUCUGGGCCUAAUUAGCAGAGAUCG ...(((((--------(((..((....))..).))))))).((((((....))))))..............((((((((.......))))))))((((((...........))))))... ( -32.60) >DroSec_CAF1 17435 112 + 1 AUCAGGAC--------AUCAAUCAAAUGGCCGCAUGUCCUUUGGCCUGGUCGGGCCAUUUAAUUUCGCAUUUGUGCCAUUUAUUUUAUGGCACAUUUCUGGGCCUAAUUAGCAGGGAUCG ...(((((--------(((..((....))..).))))))).((((((....)))))).((((((..((...((((((((.......)))))))).......))..))))))......... ( -32.20) >DroEre_CAF1 15397 111 + 1 AUCGGGAC--------AUCAAUCAAAUGGCCGCAUGUCCUUUGGCCUGGUCG-GCCAUUUAAUUUCGCAUUUGUGCCAUUUAUUUUAUGGCACAUUUCUGGUCCUAAUUAGCAGGGAGCG ...(((((--------.......((((((((((..(((....)))...).))-))))))).....((.(..((((((((.......))))))))..).))))))).....((.....)). ( -36.00) >DroYak_CAF1 15881 120 + 1 AUCGGGACAUCAGGACAUCAAUCAAAUGGCCCCAUGUCCUUUGGCCUGGUCGGGCCAUUUAAUUUCGCAUUUGUGCCAUUUAUUUUAUGGCACAUUUCUGGGCCUAAUUAGCAGGGUUCG ...(((((((..((.(((.......))).))..))))))).((((((....))))))..............((((((((.......)))))))).....((((((........)))))). ( -39.70) >consensus AUCAGGAC________AUCAAUCAAAUGGCCGCAUGUCCUUUGGCCUGGUCGGGCCAUUUAAUUUCGCAUUUGUGCCAUUUAUUUUAUGGCACAUUUCUGGGCCUAAUUAGCAGGGAUCG ..(((((................((((((((...((.((........)).))))))))))...........((((((((.......)))))))).)))))(((((........))).)). (-28.77 = -28.65 + -0.12)


| Location | 4,631,011 – 4,631,123 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.39 |
| Mean single sequence MFE | -35.80 |
| Consensus MFE | -28.73 |
| Energy contribution | -29.48 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.07 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.918581 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4631011 112 - 22224390 CGAUCUCUGCUAAUUAGGCCCAGAAAUGUGCCAUAAAAUAAAUGGCACAAAUGCGAAAUUAAAUGGCCCGACCAGGCCAAAGGACAUGCGGCCAUUUGAUUGAU--------GUCCUGAU ............((((((........((((((((.......))))))))...(((.((((((((((((((.((........))...)).))))))))))))..)--------)))))))) ( -33.20) >DroSec_CAF1 17435 112 - 1 CGAUCCCUGCUAAUUAGGCCCAGAAAUGUGCCAUAAAAUAAAUGGCACAAAUGCGAAAUUAAAUGGCCCGACCAGGCCAAAGGACAUGCGGCCAUUUGAUUGAU--------GUCCUGAU ............((((((........((((((((.......))))))))...(((.((((((((((((((.((........))...)).))))))))))))..)--------)))))))) ( -33.20) >DroEre_CAF1 15397 111 - 1 CGCUCCCUGCUAAUUAGGACCAGAAAUGUGCCAUAAAAUAAAUGGCACAAAUGCGAAAUUAAAUGGC-CGACCAGGCCAAAGGACAUGCGGCCAUUUGAUUGAU--------GUCCCGAU .((.....))..((..((((......((((((((.......)))))))).......(((((((((((-((......((...)).....)))))))))))))...--------))))..)) ( -37.60) >DroYak_CAF1 15881 120 - 1 CGAACCCUGCUAAUUAGGCCCAGAAAUGUGCCAUAAAAUAAAUGGCACAAAUGCGAAAUUAAAUGGCCCGACCAGGCCAAAGGACAUGGGGCCAUUUGAUUGAUGUCCUGAUGUCCCGAU .....((((.....))))........((((((((.......))))))))....((........(((((......)))))..((((((.(((.((((.....)))).))).)))))))).. ( -39.20) >consensus CGAUCCCUGCUAAUUAGGCCCAGAAAUGUGCCAUAAAAUAAAUGGCACAAAUGCGAAAUUAAAUGGCCCGACCAGGCCAAAGGACAUGCGGCCAUUUGAUUGAU________GUCCCGAU ............((((((........((((((((.......)))))))).......((((((((((((((.((........))...)).)))))))))))).............)))))) (-28.73 = -29.48 + 0.75)



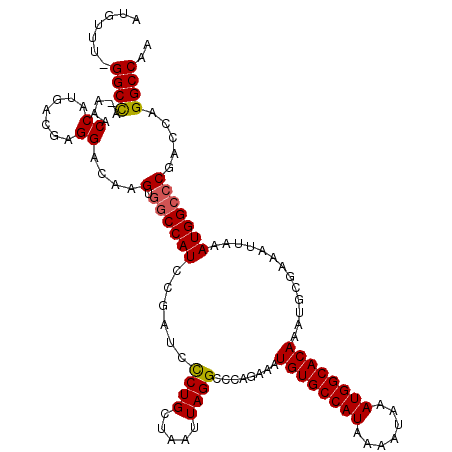
| Location | 4,631,043 – 4,631,161 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.22 |
| Mean single sequence MFE | -40.48 |
| Consensus MFE | -34.21 |
| Energy contribution | -35.40 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.24 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.889339 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4631043 118 + 22224390 UUGGCCUGGUCGGGCCAUUUAAUUUCGCAUUUGUGCCAUUUAUUUUAUGGCACAUUUCUGGGCCUAAUUAGCAGAGAUCGGAUGGCCACUUGUCCUCGUCAUGGUUU-UGGCC-AAACAU ((((((......(((((((..(((((.....((((((((.......)))))))).......((.......)).)))))..)))))))......((.......))...-.))))-)).... ( -41.10) >DroSec_CAF1 17467 118 + 1 UUGGCCUGGUCGGGCCAUUUAAUUUCGCAUUUGUGCCAUUUAUUUUAUGGCACAUUUCUGGGCCUAAUUAGCAGGGAUCGGAUGGCCACUUGUCCUCGUCAUGGUUU-UGGCC-AAACAU ((((((......(((((((..((..(.....((((((((.......)))))))).......((.......)).)..))..)))))))......((.......))...-.))))-)).... ( -40.30) >DroEre_CAF1 15429 117 + 1 UUGGCCUGGUCG-GCCAUUUAAUUUCGCAUUUGUGCCAUUUAUUUUAUGGCACAUUUCUGGUCCUAAUUAGCAGGGAGCGGAUGGUCACUUGUCCUCGUCAUGGUUU-UGGCC-AAACAU ((((((.(((.(-((((..............((((((((.......))))))))..((((.((((........)))).))))))))))))...((.......))...-.))))-)).... ( -39.00) >DroYak_CAF1 15921 120 + 1 UUGGCCUGGUCGGGCCAUUUAAUUUCGCAUUUGUGCCAUUUAUUUUAUGGCACAUUUCUGGGCCUAAUUAGCAGGGUUCGGAUGGCCACUUGUCCUCGUCAUGGUUUCUAGCCAAAACAU .((((..((.(.(((((..............((((((((.......))))))))..(((((((((........))))))))))))))....).))..))))((((.....))))...... ( -41.50) >consensus UUGGCCUGGUCGGGCCAUUUAAUUUCGCAUUUGUGCCAUUUAUUUUAUGGCACAUUUCUGGGCCUAAUUAGCAGGGAUCGGAUGGCCACUUGUCCUCGUCAUGGUUU_UGGCC_AAACAU .((((((....))))))..............((((((((.......))))))))..(((((((((........))))))))).(((((.....((.......))....)))))....... (-34.21 = -35.40 + 1.19)


| Location | 4,631,043 – 4,631,161 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.22 |
| Mean single sequence MFE | -32.95 |
| Consensus MFE | -28.90 |
| Energy contribution | -29.02 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.918496 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4631043 118 - 22224390 AUGUUU-GGCCA-AAACCAUGACGAGGACAAGUGGCCAUCCGAUCUCUGCUAAUUAGGCCCAGAAAUGUGCCAUAAAAUAAAUGGCACAAAUGCGAAAUUAAAUGGCCCGACCAGGCCAA .((..(-(((((-...((.......)).....))))))..)).(((..(((.....)))..)))..((((((((.......))))))))..............(((((......))))). ( -35.60) >DroSec_CAF1 17467 118 - 1 AUGUUU-GGCCA-AAACCAUGACGAGGACAAGUGGCCAUCCGAUCCCUGCUAAUUAGGCCCAGAAAUGUGCCAUAAAAUAAAUGGCACAAAUGCGAAAUUAAAUGGCCCGACCAGGCCAA .((..(-(((((-...((.......)).....))))))..))...((((.....))))........((((((((.......))))))))..............(((((......))))). ( -33.90) >DroEre_CAF1 15429 117 - 1 AUGUUU-GGCCA-AAACCAUGACGAGGACAAGUGACCAUCCGCUCCCUGCUAAUUAGGACCAGAAAUGUGCCAUAAAAUAAAUGGCACAAAUGCGAAAUUAAAUGGC-CGACCAGGCCAA ...(((-((((.-...((.......))...((((......))))............)).)))))..((((((((.......))))))))..............((((-(.....))))). ( -31.30) >DroYak_CAF1 15921 120 - 1 AUGUUUUGGCUAGAAACCAUGACGAGGACAAGUGGCCAUCCGAACCCUGCUAAUUAGGCCCAGAAAUGUGCCAUAAAAUAAAUGGCACAAAUGCGAAAUUAAAUGGCCCGACCAGGCCAA .....((((((.....((.......))....(.((((((......((((.....))))........((((((((.......)))))))).............))))))).....)))))) ( -31.00) >consensus AUGUUU_GGCCA_AAACCAUGACGAGGACAAGUGGCCAUCCGAUCCCUGCUAAUUAGGCCCAGAAAUGUGCCAUAAAAUAAAUGGCACAAAUGCGAAAUUAAAUGGCCCGACCAGGCCAA .......((((.....((.......))....(.((((((......((((.....))))........((((((((.......)))))))).............))))))).....)))).. (-28.90 = -29.02 + 0.13)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:14:58 2006