| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,630,790 – 4,630,898 |
| Length | 108 |
| Max. P | 0.550594 |

| Location | 4,630,790 – 4,630,898 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 65.09 |
| Mean single sequence MFE | -27.00 |
| Consensus MFE | -8.39 |
| Energy contribution | -8.89 |
| Covariance contribution | 0.51 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.31 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.550594 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
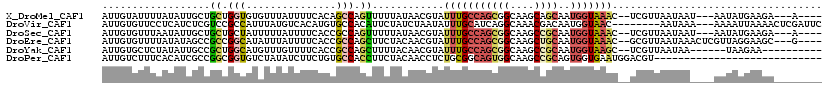
>X_DroMel_CAF1 4630790 108 + 22224390 AUUGUAUUUUAUAUUGCUGCUGGUGUGUUUAUUUUCACAGCCAGUUUUUAUAACGUAUUUGCCAGCGGCAAGCAGCAAUGGUAAAC--UCGUUAAUAAU---AAUAUGAAGA---A---- ......((((((((((..(((((((((........)))).))))).....(((((..(((((((...((.....))..))))))).--.)))))....)---))))))))).---.---- ( -31.70) >DroVir_CAF1 109431 109 + 1 AUUGUGUUCCUCAUCUCGUCCGCCAUUUAUGUCACAUGUGCCACAUUCUAUCUAAUAUUUGCAUCAGGCAAACGACAAUGGUAAC--------AAUAAA---AAAAUUAAAACUCGAUUC (((((...((.....((((..(((....(((.((.((((...............)))).)))))..)))..))))....))..))--------)))...---.................. ( -14.96) >DroSec_CAF1 17214 108 + 1 AUUGUGUUUAAUAUUGCUGCUGCUAUUUUUAUUUUCACCGCCAGUUUUUAUAACGUAUUUGCCAGCGGCAAGCCGCAAUGGUAAAC--UCGUUAAUAAU---AAUAUGAAGA---A---- ...(((...((((..((....))......))))..))).....(((.((((((((..(((((((((((....))))..))))))).--.)))).)))).---))).......---.---- ( -23.00) >DroEre_CAF1 15172 111 + 1 AUUGUGUUUUAUAUAGCCGCCGGCAUAUUUAUUUUCACCGCCAGCUUCUACAACGUAUUUGCCAGCGGCAAGCUGCAAUGGUAAAC--GCGUUAAUAAACUCGUUAGGAAGC---G---- .....(((((...((((.((.(((...............))).))......((((..(((((((((((....))))..))))))).--.)))).........)))).)))))---.---- ( -30.16) >DroYak_CAF1 15674 102 + 1 AUUGUGCUCUAUAUUGCCGCUGGCAUGUUUGUUUUCACCGCCAGCUUUUACAACGUAUUUGCCAGCGGCAAGCCGCAAUGGUAAGC--UCGUUAAUAA------UAAGAA---------- .......(((.(((((..((((((.((........))..))))))......((((..(((((((((((....))))..))))))).--.))))..)))------))))).---------- ( -31.40) >DroPer_CAF1 51801 92 + 1 AUUGUCUUUCACAUCGCCGGCGGUGUCUAUAUCUUCUGUGCCACCUUCUACAACCUCUGCGGCAGUGGCAAGCCGCAGUGGUGAAUGGACGU---------------------------- ...(((((((((((((....))))))..........................(((.(((((((........))))))).)))))).))))..---------------------------- ( -30.80) >consensus AUUGUGUUUUAUAUUGCCGCCGGCAUGUUUAUUUUCACCGCCAGCUUCUACAACGUAUUUGCCAGCGGCAAGCCGCAAUGGUAAAC__UCGUUAAUAA____AAUAUGAAGA________ ..................((.(((...............))).))............(((((((((((....))))..)))))))................................... ( -8.39 = -8.89 + 0.51)

| Location | 4,630,790 – 4,630,898 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 65.09 |
| Mean single sequence MFE | -26.95 |
| Consensus MFE | -7.77 |
| Energy contribution | -8.13 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.56 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.29 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.512552 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4630790 108 - 22224390 ----U---UCUUCAUAUU---AUUAUUAACGA--GUUUACCAUUGCUGCUUGCCGCUGGCAAAUACGUUAUAAAAACUGGCUGUGAAAAUAAACACACCAGCAGCAAUAUAAAAUACAAU ----.---..........---.........(.--((((...(((((((((((((...)))))...............(((.((((........)))))))))))))))...)))).)... ( -23.10) >DroVir_CAF1 109431 109 - 1 GAAUCGAGUUUUAAUUUU---UUUAUU--------GUUACCAUUGUCGUUUGCCUGAUGCAAAUAUUAGAUAGAAUGUGGCACAUGUGACAUAAAUGGCGGACGAGAUGAGGAACACAAU ..................---.....(--------(((.((..(.((((((((((((((....)))))......((((.((....)).))))....))))))))).)...)))))).... ( -23.20) >DroSec_CAF1 17214 108 - 1 ----U---UCUUCAUAUU---AUUAUUAACGA--GUUUACCAUUGCGGCUUGCCGCUGGCAAAUACGUUAUAAAAACUGGCGGUGAAAAUAAAAAUAGCAGCAGCAAUAUUAAACACAAU ----.---..........---...........--(((((..(((((.(((..((((..(.................)..))))..)..............)).)))))..)))))..... ( -20.66) >DroEre_CAF1 15172 111 - 1 ----C---GCUUCCUAACGAGUUUAUUAACGC--GUUUACCAUUGCAGCUUGCCGCUGGCAAAUACGUUGUAGAAGCUGGCGGUGAAAAUAAAUAUGCCGGCGGCUAUAUAAAACACAAU ----.---((.....((((.(((....))).)--))).......)).....((((((((((....((((((....)).))))((....)).....))))))))))............... ( -29.80) >DroYak_CAF1 15674 102 - 1 ----------UUCUUA------UUAUUAACGA--GCUUACCAUUGCGGCUUGCCGCUGGCAAAUACGUUGUAAAAGCUGGCGGUGAAAACAAACAUGCCAGCGGCAAUAUAGAGCACAAU ----------......------..........--((((.((.....)).((((((((((((....((((((....)).))))((....)).....))))))))))))....))))..... ( -32.80) >DroPer_CAF1 51801 92 - 1 ----------------------------ACGUCCAUUCACCACUGCGGCUUGCCACUGCCGCAGAGGUUGUAGAAGGUGGCACAGAAGAUAUAGACACCGCCGGCGAUGUGAAAGACAAU ----------------------------..(((..(((((..(((((((........)))))))..(((((....(((((.................))))).)))))))))).)))... ( -32.13) >consensus ________UCUUCAUAUU____UUAUUAACGA__GUUUACCAUUGCGGCUUGCCGCUGGCAAAUACGUUAUAAAAGCUGGCGGUGAAAAUAAAAAUACCAGCGGCAAUAUAAAACACAAU ..........................................(((((((((((.....))))....)))))))..(((((.................))))).................. ( -7.77 = -8.13 + 0.36)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:14:51 2006