
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,624,931 – 4,625,086 |
| Length | 155 |
| Max. P | 0.997960 |

| Location | 4,624,931 – 4,625,031 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 75.75 |
| Mean single sequence MFE | -24.85 |
| Consensus MFE | -14.96 |
| Energy contribution | -15.20 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.825150 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4624931 100 + 22224390 CCAAG----CCCAUCCAGGACCAACCAUUU-----GGCCAAUCCCGCACCGUCU-------G-UCAUUAACGUAAUUCAAUAACCUUCAUCUGGCCAGUUGGCCAUGAUGAUUGUUG ....(----(.......((.((((....))-----))))......)).......-------.-.................((((..((((((((((....))))).)))))..)))) ( -23.32) >DroSec_CAF1 11462 91 + 1 -------------CCCAGGACCUCCCAUUU-----GGCCAAUCCCGCACCGUCU-------G-UCAUUAACGUAAUUCAAUAACCUUCAUCUGGCCAGUUGGCCAUGAUGAUUGUUG -------------....((.((........-----))))...............-------.-.................((((..((((((((((....))))).)))))..)))) ( -22.00) >DroEre_CAF1 9431 100 + 1 CCAGG----CUCUACCAGGUCCUCCCAUUU-----GGCCAAUCCCUCACCGUCU-------G-CCAUUACUGUAAUUCAAUAACCUUCAUCUGGCCAGUUGGCCAUGAUGAUGGUUG .((((----(.......((.....((....-----)).....))......))))-------)-.................(((((.((((((((((....))))).))))).))))) ( -27.32) >DroYak_CAF1 9817 107 + 1 GCAGG----CUCUGGCAGGACCUCCCAUUC-----GGUCAAUCCCGUACCGUCUGUCGCCUG-UCAUUAAUGUAAUUCAAUAACCUUCAUCUCGCCAGUUGGCCGUGAUGAUUGUUG (((((----(...(((((((((........-----)))).....((...)).))))))))))-)................((((..(((((.((((....)).)).)))))..)))) ( -30.50) >DroAna_CAF1 72772 106 + 1 CCACUUUCUCCCUUCUCCCACCCGCCACUUUCUCCGGCCAACC----GUCGUCC-------GACCAUUAAUGUAAUUUAAUAACCUCCAUCUGGCCAGUCGGCCAUGAUGAUUGUUG .......................(((.........)))((((.----((((((.-------....(((((......)))))..........(((((....))))).)))))).)))) ( -21.10) >consensus CCAGG____CCCUUCCAGGACCUCCCAUUU_____GGCCAAUCCCGCACCGUCU_______G_UCAUUAAUGUAAUUCAAUAACCUUCAUCUGGCCAGUUGGCCAUGAUGAUUGUUG ................................................................................((((..((((((((((....))))).)))))..)))) (-14.96 = -15.20 + 0.24)



| Location | 4,624,931 – 4,625,031 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 75.75 |
| Mean single sequence MFE | -32.96 |
| Consensus MFE | -19.26 |
| Energy contribution | -20.58 |
| Covariance contribution | 1.32 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.984334 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4624931 100 - 22224390 CAACAAUCAUCAUGGCCAACUGGCCAGAUGAAGGUUAUUGAAUUACGUUAAUGA-C-------AGACGGUGCGGGAUUGGCC-----AAAUGGUUGGUCCUGGAUGGG----CUUGG ......(((((.(((((....))))))))))..((((((((......)))))))-)-------...(.((.((((((..(((-----....)))..)))))).)).).----..... ( -41.30) >DroSec_CAF1 11462 91 - 1 CAACAAUCAUCAUGGCCAACUGGCCAGAUGAAGGUUAUUGAAUUACGUUAAUGA-C-------AGACGGUGCGGGAUUGGCC-----AAAUGGGAGGUCCUGGG------------- ......(((((.(((((....))))))))))..((((((((......)))))))-)-------........(((((((..((-----....))..)))))))..------------- ( -32.00) >DroEre_CAF1 9431 100 - 1 CAACCAUCAUCAUGGCCAACUGGCCAGAUGAAGGUUAUUGAAUUACAGUAAUGG-C-------AGACGGUGAGGGAUUGGCC-----AAAUGGGAGGACCUGGUAGAG----CCUGG ..(((.(((((.(((((....))))))))))..(((((((.....)))))))..-.-------....)))..(((....(((-----(...((.....))))))....----))).. ( -31.80) >DroYak_CAF1 9817 107 - 1 CAACAAUCAUCACGGCCAACUGGCGAGAUGAAGGUUAUUGAAUUACAUUAAUGA-CAGGCGACAGACGGUACGGGAUUGACC-----GAAUGGGAGGUCCUGCCAGAG----CCUGC ......(((((.((.((....)))).)))))..((((((((......)))))))-)(((((.....)(((..((((((..((-----....))..)))))))))...)----))).. ( -32.60) >DroAna_CAF1 72772 106 - 1 CAACAAUCAUCAUGGCCGACUGGCCAGAUGGAGGUUAUUAAAUUACAUUAAUGGUC-------GGACGAC----GGUUGGCCGGAGAAAGUGGCGGGUGGGAGAAGGGAGAAAGUGG ...((.((..(....((.(((.((((......(..((((((......))))))..)-------...(..(----((....)))..)....)))).))).)).....)..))...)). ( -27.10) >consensus CAACAAUCAUCAUGGCCAACUGGCCAGAUGAAGGUUAUUGAAUUACAUUAAUGA_C_______AGACGGUGCGGGAUUGGCC_____AAAUGGGAGGUCCUGGAAGAG____CCUGG .(((..(((((.(((((....))))))))))..)))...................................(((((((..((.........))..)))))))............... (-19.26 = -20.58 + 1.32)

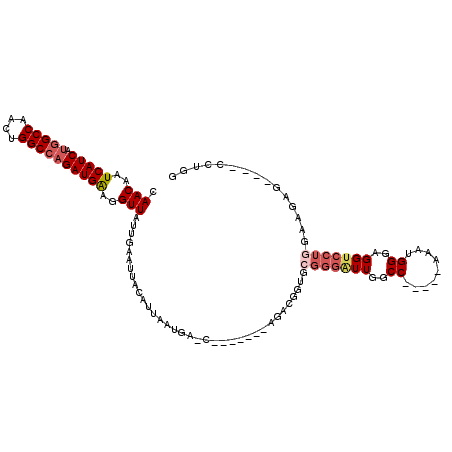
| Location | 4,624,992 – 4,625,086 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 73.07 |
| Mean single sequence MFE | -28.20 |
| Consensus MFE | -13.54 |
| Energy contribution | -14.85 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.97 |
| Structure conservation index | 0.48 |
| SVM decision value | 2.97 |
| SVM RNA-class probability | 0.997960 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4624992 94 + 22224390 AAUAACCU-UCAUCUGGCCAGUUGGCCAUGAUGAUUGUUGAUUGUUUUUCAUGCU------AUGCCAGAGCGCCUCGAA-----AUUUGCAAUUAAUAAUUG------GUAC ....(((.-((((((((((....))))).)))))(((((((((((.((((.((((------.......))))....)))-----)...)))))))))))..)------)).. ( -30.10) >DroPse_CAF1 41463 106 + 1 AAUAACCCAGCAUCUGGACAGUUGGAUA---UGAUUGCUGAUUGUUUUUCAUUCGCCAGAGCCGCCAGAGCGCUUC---GAGUGGGGUGCAAUUAAUGAUAUUCGAGUGCAC ......((((...)))).((.(((((((---(.((...(((((((.(..((((((...((((.((....)))))))---)))))..).))))))))).)))))))).))... ( -33.00) >DroSec_CAF1 11514 94 + 1 AAUAACCU-UCAUCUGGCCAGUUGGCCAUGAUGAUUGUUGAUUGUUUUUCAUGCG------AUGCCAGAGCGCCUCGAA-----AUUUGCAAUUAAUAAUUG------GCAC ........-((((((((((....))))).))))).(((..((((((.....((((------(.....(((...)))...-----..)))))...))))))..------))). ( -29.50) >DroEre_CAF1 9492 87 + 1 AAUAACCU-UCAUCUGGCCAGUUGGCCAUGAUGAUGGUUGAUUGUUUUUCACGCG------AUGCCAGAGCGCCUCGAA------------AUUAAUAAUUG------CCAA ...((((.-((((((((((....))))).))))).))))(((((((.......((------(.((......)).)))..------------...))))))).------.... ( -27.30) >DroYak_CAF1 9885 94 + 1 AAUAACCU-UCAUCUCGCCAGUUGGCCGUGAUGAUUGUUGAUUGUUUUUUAUGCG------AUGCCAUAGCGUCUCGCA-----AUUUGCAAUUAAUAAUUG------GCAC .....((.-(((((.((((....)).)).)))))(((((((((((...((.((.(------((((....))))).)).)-----)...)))))))))))..)------)... ( -27.00) >DroAna_CAF1 72839 75 + 1 AAUAACCU-CCAUCUGGCCAGUCGGCCAUGAUGAUUGUUGAUUGUUUUACG-------------------------AAA-----AUAUGCAAUUAAUAAUUG------GCAC .....((.-.(((((((((....))))).))))((((((((((((......-------------------------...-----....)))))))))))).)------)... ( -22.32) >consensus AAUAACCU_UCAUCUGGCCAGUUGGCCAUGAUGAUUGUUGAUUGUUUUUCAUGCG______AUGCCAGAGCGCCUCGAA_____AUUUGCAAUUAAUAAUUG______GCAC ..........(((((((((....))))).))))((((((((((((...........................................))))))))))))............ (-13.54 = -14.85 + 1.31)



| Location | 4,624,992 – 4,625,086 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 73.07 |
| Mean single sequence MFE | -22.08 |
| Consensus MFE | -10.95 |
| Energy contribution | -12.12 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.50 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4624992 94 - 22224390 GUAC------CAAUUAUUAAUUGCAAAU-----UUCGAGGCGCUCUGGCAU------AGCAUGAAAAACAAUCAACAAUCAUCAUGGCCAACUGGCCAGAUGA-AGGUUAUU ..((------(................(-----((((..((((....))..------.)).)))))............(((((.(((((....))))))))))-.))).... ( -21.40) >DroPse_CAF1 41463 106 - 1 GUGCACUCGAAUAUCAUUAAUUGCACCCCACUC---GAAGCGCUCUGGCGGCUCUGGCGAAUGAAAAACAAUCAGCAAUCA---UAUCCAACUGUCCAGAUGCUGGGUUAUU (((((.(..(......)..).)))))((((...---...(((.((((((((...(((...((((..............)))---)..))).))).))))))))))))..... ( -22.44) >DroSec_CAF1 11514 94 - 1 GUGC------CAAUUAUUAAUUGCAAAU-----UUCGAGGCGCUCUGGCAU------CGCAUGAAAAACAAUCAACAAUCAUCAUGGCCAACUGGCCAGAUGA-AGGUUAUU ((((------((((.....)))).....-----...((.((......)).)------)))))...........(((..(((((.(((((....))))))))))-..)))... ( -23.20) >DroEre_CAF1 9492 87 - 1 UUGG------CAAUUAUUAAU------------UUCGAGGCGCUCUGGCAU------CGCGUGAAAAACAAUCAACCAUCAUCAUGGCCAACUGGCCAGAUGA-AGGUUAUU ....------..........(------------((((.(..((....))..------)...))))).......((((.(((((.(((((....))))))))))-.))))... ( -25.60) >DroYak_CAF1 9885 94 - 1 GUGC------CAAUUAUUAAUUGCAAAU-----UGCGAGACGCUAUGGCAU------CGCAUAAAAAACAAUCAACAAUCAUCACGGCCAACUGGCGAGAUGA-AGGUUAUU ((((------((........((((....-----.)))).......))))))------................(((..(((((.((.((....)))).)))))-..)))... ( -22.16) >DroAna_CAF1 72839 75 - 1 GUGC------CAAUUAUUAAUUGCAUAU-----UUU-------------------------CGUAAAACAAUCAACAAUCAUCAUGGCCGACUGGCCAGAUGG-AGGUUAUU ((((------.(((.....)))))))..-----...-------------------------............(((..(((((.(((((....))))))))))-..)))... ( -17.70) >consensus GUGC______CAAUUAUUAAUUGCAAAU_____UUCGAGGCGCUCUGGCAU______CGCAUGAAAAACAAUCAACAAUCAUCAUGGCCAACUGGCCAGAUGA_AGGUUAUU .........................................((....))........................(((..(((((.(((((....))))))))))...)))... (-10.95 = -12.12 + 1.17)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:14:46 2006