| Sequence ID | X_DroMel_CAF1 |
|---|---|
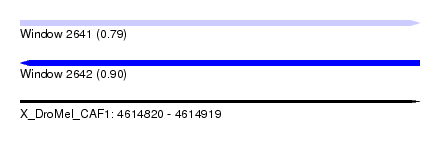
| Location | 4,614,820 – 4,614,919 |
| Length | 99 |
| Max. P | 0.900078 |

| Location | 4,614,820 – 4,614,919 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 59.42 |
| Mean single sequence MFE | -26.96 |
| Consensus MFE | -13.02 |
| Energy contribution | -14.28 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.790487 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4614820 99 + 22224390 ---------------------GACUGUUUACCAUUAAGAUCUUCUUGCGAUUCCAUUUUUCUGUCCACAAGUGCCCAGCAGAGGAAUGUUGUCCUCUUCACCAGCACCAUAAUGGGCAGA ---------------------.............((((.....))))............((((((((...((((...(.((((((......)))))).)....)))).....)))))))) ( -23.50) >DroSec_CAF1 463 92 + 1 ----------------------------GACCACCAAGAUCUACGUCCGAUUCGAUAUUCCUGUCCACAAAUGCCCACGAGAGGAAAGUAAUCCUCUUCACCAGCACCAUGAUGGCCAGA ----------------------------.........((((.......))))........(((.(((((..(((....(((((((......))))))).....)))...)).))).))). ( -18.00) >DroEre_CAF1 447 111 + 1 CUGCUGGAUUCCUGGUAAAGCGACUGUGU---------AUCCUCGUCCGACUCCACAUUCCUGUCGGGCAAUACCCAGUCCAGGAAACCAGUCUUCCUCACCAGCACCAUGACCGGCAGA (((((((..((.((((...(((((((.((---------((....(((((((...........))))))).)))).))))).(((((.......))))).....)))))).))))))))). ( -41.00) >DroYak_CAF1 431 111 + 1 GUGCUGCAUUUCUGGUAAAGCGACUCUUU---------AUACUCAUUCGCUUCCUUAUUCCUGUCUAGCAAUGCCCAGUCAAAGAAUGUGGUCCUUCGCACCAGCACCAUGACGGGCAGA .(((((((.....((..((((((......---------........)))))))).......)))..)))).(((((.((((.....((((((.......)))).))...))))))))).. ( -25.34) >consensus _____________________GACUGUUU_________AUCCUCGUCCGAUUCCAUAUUCCUGUCCACAAAUGCCCAGUAGAGGAAAGUAGUCCUCCUCACCAGCACCAUGACGGGCAGA ............................................................(((((((....(((...((((((((......))))))))....)))......))))))). (-13.02 = -14.28 + 1.25)

| Location | 4,614,820 – 4,614,919 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 59.42 |
| Mean single sequence MFE | -34.02 |
| Consensus MFE | -16.60 |
| Energy contribution | -16.73 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.49 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.900078 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
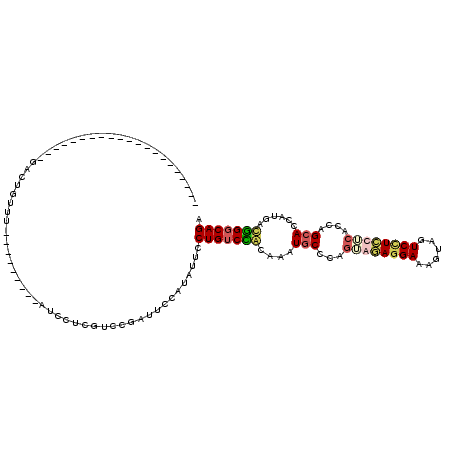
>X_DroMel_CAF1 4614820 99 - 22224390 UCUGCCCAUUAUGGUGCUGGUGAAGAGGACAACAUUCCUCUGCUGGGCACUUGUGGACAGAAAAAUGGAAUCGCAAGAAGAUCUUAAUGGUAAACAGUC--------------------- ((((.((((...((((((.(...((((((......))))))..).)))))).)))).)))).........((....)).....................--------------------- ( -30.00) >DroSec_CAF1 463 92 - 1 UCUGGCCAUCAUGGUGCUGGUGAAGAGGAUUACUUUCCUCUCGUGGGCAUUUGUGGACAGGAAUAUCGAAUCGGACGUAGAUCUUGGUGGUC---------------------------- ((((.((((...((((((..(((.(((((......))))))))..)))))).)))).))))..(((((((((.......))).))))))...---------------------------- ( -29.90) >DroEre_CAF1 447 111 - 1 UCUGCCGGUCAUGGUGCUGGUGAGGAAGACUGGUUUCCUGGACUGGGUAUUGCCCGACAGGAAUGUGGAGUCGGACGAGGAU---------ACACAGUCGCUUUACCAGGAAUCCAGCAG .((((.((......(.(((((((((..(((((..((((((...((((.....)))).))))))((((...((....))...)---------)))))))).))))))))).)..)).)))) ( -42.90) >DroYak_CAF1 431 111 - 1 UCUGCCCGUCAUGGUGCUGGUGCGAAGGACCACAUUCUUUGACUGGGCAUUGCUAGACAGGAAUAAGGAAGCGAAUGAGUAU---------AAAGAGUCGCUUUACCAGAAAUGCAGCAC ..(((((((((.((((.((((.(....)))))))))...)))).))))).((((.(.((........(((((((.(......---------..)...)))))))........))))))). ( -33.29) >consensus UCUGCCCAUCAUGGUGCUGGUGAAGAGGACUACAUUCCUCGACUGGGCAUUGCUGGACAGGAAUAUGGAAUCGGACGAAGAU_________AAACAGUC_____________________ ((((.((((...((((((.(...((((((......))))))..).)))))).)))).))))........................................................... (-16.60 = -16.73 + 0.12)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:14:43 2006