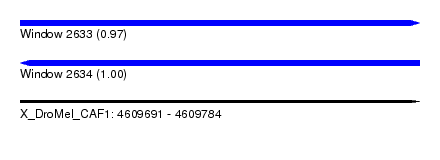
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,609,691 – 4,609,784 |
| Length | 93 |
| Max. P | 0.997792 |

| Location | 4,609,691 – 4,609,784 |
|---|---|
| Length | 93 |
| Sequences | 3 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 78.02 |
| Mean single sequence MFE | -23.65 |
| Consensus MFE | -17.55 |
| Energy contribution | -18.33 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.965683 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4609691 93 + 22224390 AUAGGUU-GAGCGGUUAUGUCAGAGGUGAGCAUACAUACAUACAUACCUCUCACUUUCGCACACCUCUGCCCUCCCUCUCCCACGCUUCCGCGC ...((..-(((.((....(.((((((((.((...........................)).)))))))))...)))))..)).(((....))). ( -25.73) >DroSec_CAF1 33835 85 + 1 AUAGGAU-GAAAGGUUAUGUCAGAGGUGAGCAUAC--------AUACCUCUCGCUUUCGCACACCUCUGCCCUCCCUCUCCCUCACUCGCGCGC ..(((..-((.(((....(.((((((((.((....--------...............)).)))))))))....))).)))))........... ( -19.41) >DroEre_CAF1 36973 82 + 1 AUAGGGAAGAGGGGUUAAGUCAGAGGUGAACACAC--------AUACCUCUCGCUCUCGCACACCUCUGCCCUCCCUUUCUA----UCCUGCGC ..((((.(((((.((..(((.(((((((.......--------.))))))).)))...))...))))).)))).........----........ ( -25.80) >consensus AUAGGAU_GAGAGGUUAUGUCAGAGGUGAGCAUAC________AUACCUCUCGCUUUCGCACACCUCUGCCCUCCCUCUCCC_C_CUCCCGCGC ........((((((....(.((((((((.((...........................)).)))))))))....)))))).............. (-17.55 = -18.33 + 0.78)

| Location | 4,609,691 – 4,609,784 |
|---|---|
| Length | 93 |
| Sequences | 3 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 78.02 |
| Mean single sequence MFE | -33.17 |
| Consensus MFE | -22.05 |
| Energy contribution | -22.50 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.66 |
| SVM decision value | 2.93 |
| SVM RNA-class probability | 0.997792 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4609691 93 - 22224390 GCGCGGAAGCGUGGGAGAGGGAGGGCAGAGGUGUGCGAAAGUGAGAGGUAUGUAUGUAUGUAUGCUCACCUCUGACAUAACCGCUC-AACCUAU ((((....)))).....(((..((((.(..((((((....)).((((((..(((((....)))))..)))))).))))..).))))-..))).. ( -36.00) >DroSec_CAF1 33835 85 - 1 GCGCGCGAGUGAGGGAGAGGGAGGGCAGAGGUGUGCGAAAGCGAGAGGUAU--------GUAUGCUCACCUCUGACAUAACCUUUC-AUCCUAU .(((....))).((((..(..(((.(((((((((((....)))...((((.--------...))))))))))))......)))..)-.)))).. ( -34.70) >DroEre_CAF1 36973 82 - 1 GCGCAGGA----UAGAAAGGGAGGGCAGAGGUGUGCGAGAGCGAGAGGUAU--------GUGUGUUCACCUCUGACUUAACCCCUCUUCCCUAU ....(((.----.(((..((..(((((((((((.(((..(.(....).)..--------...))).)))))))).)))..))..)))..))).. ( -28.80) >consensus GCGCGGGAG_G_GGGAGAGGGAGGGCAGAGGUGUGCGAAAGCGAGAGGUAU________GUAUGCUCACCUCUGACAUAACCCCUC_AACCUAU .(((....)))...(((.((....((((((((..((....))(((..((((........)))).)))))))))).)....)).)))........ (-22.05 = -22.50 + 0.45)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:14:35 2006