| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,602,579 – 4,602,681 |
| Length | 102 |
| Max. P | 0.986825 |

| Location | 4,602,579 – 4,602,681 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 71.13 |
| Mean single sequence MFE | -25.73 |
| Consensus MFE | -13.29 |
| Energy contribution | -15.98 |
| Covariance contribution | 2.69 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.52 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.929092 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4602579 102 + 22224390 UACUUGUAUAUAUAUA-UUGUUGCAGGCAACUGUAGCUUAAUCCCGAAACUUCAUGUCCAGGGUGCUCAUG-UUGGGCUUGCAUCUUCCAGAUGCAAAGAUACU .....((((.......-..(((((((....)))))))..................((((((.((....)).-))))))(((((((.....)))))))..)))). ( -29.80) >DroSec_CAF1 27543 97 + 1 -----AUAUCCAUCCU-UUGUUGCAGGCAGCUGUAGCUUAAUCCCGUAACUUCAUGUCCAGGGUGCUCAUG-UUGGGCUUGCAUCUUUCGGAUGCAAAGAUACU -----.(((((((((.-..(.(((((((.((((.(((.....(((...((.....))...))).))))).)-)...))))))).)....)))))....)))).. ( -26.80) >DroEre_CAF1 30528 88 + 1 UGCCUAAAUAUAUAUUUUUUUUUCAGGCAGCUAUAGCUUUUUCCCGAAACUUCAUG----------------UUGGACUUGCAUCUUUCAGAUGCGAGGAUACA ((((((((............))).)))))((....))....(((...(((.....)----------------)))))((((((((.....))))))))...... ( -19.20) >DroYak_CAF1 29032 88 + 1 --------CAUAUA--------UCAGGCAGCUGUAGCUUAAGCCCGAAACAUCAUGCCCAGUGUGCCCAAGCUUGGGCUUGCAUCUUCCAGAUGCAAAGAUACU --------......--------((.((((((....)))...))).))...(((..((((((.((......))))))))(((((((.....))))))).)))... ( -27.10) >consensus _____AUAUAUAUAUU_UUGUUGCAGGCAGCUGUAGCUUAAUCCCGAAACUUCAUGUCCAGGGUGCUCAUG_UUGGGCUUGCAUCUUCCAGAUGCAAAGAUACU ...................(((((((....)))))))...........................(((((....)))))(((((((.....)))))))....... (-13.29 = -15.98 + 2.69)

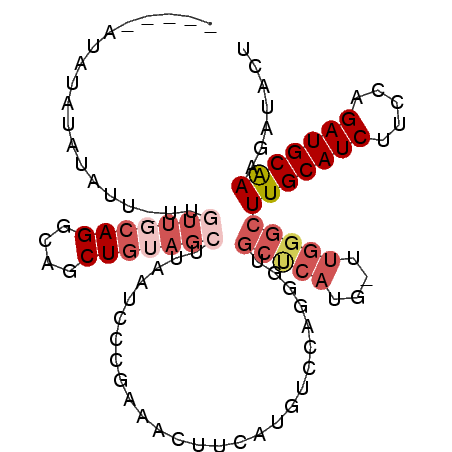

| Location | 4,602,579 – 4,602,681 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 71.13 |
| Mean single sequence MFE | -25.30 |
| Consensus MFE | -16.70 |
| Energy contribution | -18.07 |
| Covariance contribution | 1.37 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.66 |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.986825 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4602579 102 - 22224390 AGUAUCUUUGCAUCUGGAAGAUGCAAGCCCAA-CAUGAGCACCCUGGACAUGAAGUUUCGGGAUUAAGCUACAGUUGCCUGCAACAA-UAUAUAUAUACAAGUA .((((..(((((((.....)))))))((.(((-(.(((((.(((..(((.....)))..))).....))).))))))...)).....-.......))))..... ( -24.30) >DroSec_CAF1 27543 97 - 1 AGUAUCUUUGCAUCCGAAAGAUGCAAGCCCAA-CAUGAGCACCCUGGACAUGAAGUUACGGGAUUAAGCUACAGCUGCCUGCAACAA-AGGAUGGAUAU----- .((((((((((((.(....)))))))..((..-((((..(.....)..))))..(((.((((....(((....))).)))).)))..-.))..))))))----- ( -26.10) >DroEre_CAF1 30528 88 - 1 UGUAUCCUCGCAUCUGAAAGAUGCAAGUCCAA----------------CAUGAAGUUUCGGGAAAAAGCUAUAGCUGCCUGAAAAAAAAAUAUAUAUUUAGGCA .....(((.(((((.....)))))........----------------.......(((((((....(((....))).)))))))...............))).. ( -19.50) >DroYak_CAF1 29032 88 - 1 AGUAUCUUUGCAUCUGGAAGAUGCAAGCCCAAGCUUGGGCACACUGGGCAUGAUGUUUCGGGCUUAAGCUACAGCUGCCUGA--------UAUAUG-------- .......(((((((.....)))))))(((((.((....))....)))))...((((.((((((...(((....)))))))))--------.)))).-------- ( -31.30) >consensus AGUAUCUUUGCAUCUGAAAGAUGCAAGCCCAA_CAUGAGCACCCUGGACAUGAAGUUUCGGGAUUAAGCUACAGCUGCCUGAAACAA_AAUAUAUAUAU_____ ......((((((((.....))))))))...........................((((((((....(((....))).))))))))................... (-16.70 = -18.07 + 1.37)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:14:32 2006