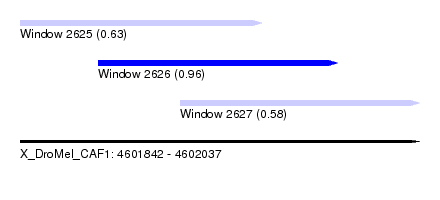
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,601,842 – 4,602,037 |
| Length | 195 |
| Max. P | 0.964679 |

| Location | 4,601,842 – 4,601,960 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.45 |
| Mean single sequence MFE | -26.41 |
| Consensus MFE | -18.75 |
| Energy contribution | -19.62 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.633612 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4601842 118 + 22224390 UAUAAUUAACGGCAAUAAAAAUCAAAGCCUGCGGAAGU--CUUCAAUUUAAGUUCGAUCUAAUUGGUCAUAAGUGAAUGACUUAUUUCCAUGGAUAUCCUUGAAUGGCGGAAGGCAUUGA ..........(((.............))).......((--((((.(((((((....(((((..(((..((((((.....))))))..))))))))...)))))))....))))))..... ( -27.22) >DroSec_CAF1 26838 118 + 1 AAUAAUUAACGGAAAUAAGAAUCAAAGCCUGCGGAAGU--CUUCAAUUUAUUUUCGAUCUAAUUGGUCAUAAGUGAAUGACUUAUUUCCAUGGAUAUCCUUGAAUGGCGGAAGGCAUUGA ..........(((((((((...........((....))--..(((.(((((((..((((.....))))..)))))))))))))))))))..((....))...((((.(....).)))).. ( -29.50) >DroEre_CAF1 29825 110 + 1 ---------CGAAUAUAGACAUCAAAGUCUGAGGAAGACCGAAGAAUU-AUGUUCGAUCUAAUUGGUCAUAAGUGAAUGACUUAUUUCCAUGGAUAUCCUUGAAUGGCGGAAGGCAUUAA ---------.......((((......))))(((((....((((.(...-.).))))(((((..(((..((((((.....))))))..)))))))).))))).((((.(....).)))).. ( -27.70) >DroYak_CAF1 28322 111 + 1 -----UU--AGAAUAUAAAAAUCAAAGUCUGAGAAAGC--CAUGAAUUAAUGUUCGAACUAAUUGGUCAUAAGUGAAUGACUUAUUGCCAUGGAUAUCCUUGAAUGACGGAAGGCAUUGA -----..--............((((.((((.......(--(((((((....)))).........(((.((((((.....)))))).)))))))...(((.........))))))).)))) ( -21.20) >consensus _____UU__CGAAAAUAAAAAUCAAAGCCUGAGGAAGU__CAUCAAUUUAUGUUCGAUCUAAUUGGUCAUAAGUGAAUGACUUAUUUCCAUGGAUAUCCUUGAAUGGCGGAAGGCAUUGA .........((((((((((.........((.....)).........))))))))))(((((..(((..((((((.....))))))..)))))))).......((((.(....).)))).. (-18.75 = -19.62 + 0.87)


| Location | 4,601,880 – 4,601,997 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 93.08 |
| Mean single sequence MFE | -33.75 |
| Consensus MFE | -30.66 |
| Energy contribution | -30.72 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.964679 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4601880 117 + 22224390 CUUCAAUUUAAGUUCGAUCUAAUUGGUCAUAAGUGAAUGACUUAUUUCCAUGGAUAUCCUUGAAUGGCGGAAGGCAUUGAAAUGGUCCUGCAAAUUGGCUCGUUUGGAUUGC--GCCAA ...........((.((((((((.((((((....((...((((.(((((((.((....)).))((((.(....).)))))))))))))...))...)))).)).)))))))).--))... ( -30.80) >DroSec_CAF1 26876 117 + 1 CUUCAAUUUAUUUUCGAUCUAAUUGGUCAUAAGUGAAUGACUUAUUUCCAUGGAUAUCCUUGAAUGGCGGAAGGCAUUGAAAUGAGCCUGCAAAUUGGCUCGUUUGGAUUGC--GCCAA ..(((.(((((((..((((.....))))..))))))))))..........(((..((((...((((.(....).)))).(((((((((........)))))))))))))...--.))). ( -35.00) >DroEre_CAF1 29856 116 + 1 GAAGAAUU-AUGUUCGAUCUAAUUGGUCAUAAGUGAAUGACUUAUUUCCAUGGAUAUCCUUGAAUGGCGGAAGGCAUUAAAAUGAGCCUGCAAAUUGGCUCGUUUGGAUUGC--GCCAA ...((((.-..))))........(((..((((((.....))))))..)))(((..((((...((((.(....).)))).(((((((((........)))))))))))))...--.))). ( -34.30) >DroYak_CAF1 28353 119 + 1 CAUGAAUUAAUGUUCGAACUAAUUGGUCAUAAGUGAAUGACUUAUUGCCAUGGAUAUCCUUGAAUGACGGAAGGCAUUGAAAUGAGCCUGCAAAUUGGCUCGUUUGGAUUGCGCGCCAA ...........((((((......((((.((((((.....)))))).)))).((....))))))))...((...(((((.(((((((((........))))))))).)).)))...)).. ( -34.90) >consensus CAUCAAUUUAUGUUCGAUCUAAUUGGUCAUAAGUGAAUGACUUAUUUCCAUGGAUAUCCUUGAAUGGCGGAAGGCAUUGAAAUGAGCCUGCAAAUUGGCUCGUUUGGAUUGC__GCCAA .......................(((..((((((.....))))))..)))(((..((((...((((.(....).)))).(((((((((........)))))))))))))......))). (-30.66 = -30.72 + 0.06)

| Location | 4,601,920 – 4,602,037 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 95.76 |
| Mean single sequence MFE | -29.60 |
| Consensus MFE | -27.41 |
| Energy contribution | -27.10 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.579864 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4601920 117 + 22224390 CUUAUUUCCAUGGAUAUCCUUGAAUGGCGGAAGGCAUUGAAAUGGUCCUGCAAAUUGGCUCGUUUGGAUUGC--GCCAACACAGUUCCCAUUAAAAUUGACUUCAAGUUAUUUACAUAG ..........((((((..(((((((((((.(....(((.(((((..((........))..))))).)))).)--))))...(((((........)))))..)))))).))))))..... ( -24.00) >DroSec_CAF1 26916 117 + 1 CUUAUUUCCAUGGAUAUCCUUGAAUGGCGGAAGGCAUUGAAAUGAGCCUGCAAAUUGGCUCGUUUGGAUUGC--GCCAACACAAUUCCCAUUAAAAUUGACUUCAAGUUAUUUACAUAG ..........((((((..(((((((((((.(....(((.(((((((((........))))))))).)))).)--))))...(((((........)))))..)))))).))))))..... ( -31.40) >DroEre_CAF1 29895 117 + 1 CUUAUUUCCAUGGAUAUCCUUGAAUGGCGGAAGGCAUUAAAAUGAGCCUGCAAAUUGGCUCGUUUGGAUUGC--GCCAACGCAAUUCCCAUUAAAAUUGACUUCAAGUUAUUUACAUAG ..........((((((..((((((((.(....).))...(((((((((........)))))))))(((((((--(....))))))))..............)))))).))))))..... ( -32.50) >DroYak_CAF1 28393 119 + 1 CUUAUUGCCAUGGAUAUCCUUGAAUGACGGAAGGCAUUGAAAUGAGCCUGCAAAUUGGCUCGUUUGGAUUGCGCGCCAACACAAUUCCCAUUGAAAUUGACUUCAAGUUAUUUACAUAG ...........((....)).((((((((.(((((((((.(((((((((........))))))))).)).))).........(((((........))))).))))..))))))))..... ( -30.50) >consensus CUUAUUUCCAUGGAUAUCCUUGAAUGGCGGAAGGCAUUGAAAUGAGCCUGCAAAUUGGCUCGUUUGGAUUGC__GCCAACACAAUUCCCAUUAAAAUUGACUUCAAGUUAUUUACAUAG ..........((((((..((((((((..((...(((((.(((((((((........))))))))).)).)))...))..))(((((........)))))..)))))).))))))..... (-27.41 = -27.10 + -0.31)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:14:29 2006