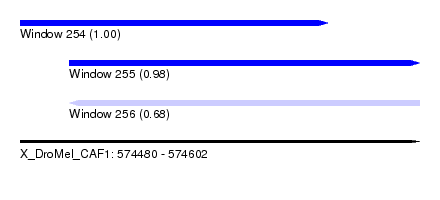
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 574,480 – 574,602 |
| Length | 122 |
| Max. P | 0.996487 |

| Location | 574,480 – 574,574 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 74.96 |
| Mean single sequence MFE | -15.30 |
| Consensus MFE | -8.40 |
| Energy contribution | -9.03 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.55 |
| SVM decision value | 2.70 |
| SVM RNA-class probability | 0.996487 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 574480 94 + 22224390 CAUCCACUUUCAAA------------------------GAAAUAAGCUCAUCAUAUCUUAUCAGUU-UUCCAAUUUUAUGUUCAUAUAUUCCACUCCGGGAAUACAUAUAUAAAAUUUA ..............------------------------............................-....((((((((((.....((((((......))))))...)))))))))).. ( -10.40) >DroSec_CAF1 38272 94 + 1 CUUUCACUUUCAAA------------------------GAAAGAAGCUCAUCAAAUCUUAGUAUUU-UUCCAAUUUUAUGUAUAUAUAUUCCCCUCCGGGAAUACAUAUAUAAAAAAUA ..............------------------------((((((.(((...........))).)))-)))..(((((.(((((((.(((((((....))))))).))))))).))))). ( -19.10) >DroSim_CAF1 37805 94 + 1 CUUUCACUUUCAAA------------------------GAAAGAAGCUCAUCAAAUCUUAGUAUUU-UCCCAAUUUUAUGUAUAUAUAUUCCACUCCGGGAAUACAUAUAUAAAAAAUA (((((.........------------------------))))).......................-.....(((((.(((((((.((((((......)))))).))))))).))))). ( -15.60) >DroEre_CAF1 38579 118 + 1 CUUCCACUUUCAAAAGGAAAAUUUGGAAUGAAUACAAAAAAAUAAGCGCAACAAAUCUUAGUAUUGUUUUCAAUUUCAUAUUCAUAUACUCCACUCCGGGAAUAUACAAGUAUA-AAUA .(((((.((((.....))))...)))))((((.((((.(...((((..........)))).).)))).))))((((.(((((..((((.(((......))).))))..))))))-))). ( -16.10) >consensus CUUCCACUUUCAAA________________________GAAAGAAGCUCAUCAAAUCUUAGUAUUU_UUCCAAUUUUAUGUACAUAUAUUCCACUCCGGGAAUACAUAUAUAAAAAAUA ..............................................................................(((((((.((((((......)))))).)))))))....... ( -8.40 = -9.03 + 0.63)

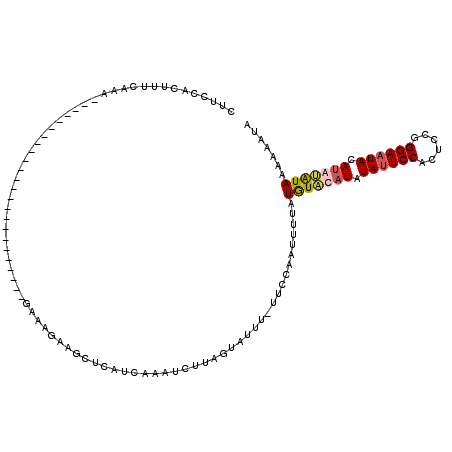
| Location | 574,495 – 574,602 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.84 |
| Mean single sequence MFE | -14.20 |
| Consensus MFE | -4.47 |
| Energy contribution | -4.47 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.31 |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.979894 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 574495 107 + 22224390 AAAUAAGCUCAUCAUAUCUUAUCAGUU-UUCCAAUUUUAUGUUCAUAUAUUCCACUCCGGGAAUACAUAUAUAAAAUUUACGUCUGUAUAUACC------------CGUUCUUUCCCACA ...................((((((..-....((((((((((.....((((((......))))))...)))))))))).....))).)))....------------.............. ( -12.90) >DroSec_CAF1 38287 111 + 1 AAAGAAGCUCAUCAAAUCUUAGUAUUU-UUCCAAUUUUAUGUAUAUAUAUUCCCCUCCGGGAAUACAUAUAUAAAAAAUACCUCUGUAUAUAUC--------CUCUCGUUCUUUCCCACA ((((((.....................-......((((((((((...(((((((....))))))).)))))))))).((((....)))).....--------......))))))...... ( -17.00) >DroSim_CAF1 37820 111 + 1 AAAGAAGCUCAUCAAAUCUUAGUAUUU-UCCCAAUUUUAUGUAUAUAUAUUCCACUCCGGGAAUACAUAUAUAAAAAAUACCUCUGUAUAUAUC--------CUCUCGUUCUUUCCCACA ((((((.....................-......((((((((((...((((((......)))))).)))))))))).((((....)))).....--------......))))))...... ( -14.40) >DroEre_CAF1 38618 109 + 1 AAAUAAGCGCAACAAAUCUUAGUAUUGUUUUCAAUUUCAUAUUCAUAUACUCCACUCCGGGAAUAUACAAGUAUA-AAUAACACUGUGUAC--C--------CUCUCGCUUCUUCCCACA ....(((((..........((((.((((((........(((((..((((.(((......))).))))..))))))-))))).)))).....--.--------....)))))......... ( -14.35) >DroYak_CAF1 41916 102 + 1 UAAUA----------AUCCAAGUAUGG-UUCCAAUUUCAUUUUCAUACACUCCACUCCGGGAAUAU------AUA-AAUAACACUGUAUGCAACCUCUCGUUCUCUCGCCCAUUCCCACA .....----------......((((((-..............))))))..........((((((((------(((-........))))).........((......))...))))))... ( -12.34) >consensus AAAUAAGCUCAUCAAAUCUUAGUAUUU_UUCCAAUUUUAUGUUCAUAUAUUCCACUCCGGGAAUACAUAUAUAAAAAAUACCUCUGUAUAUAUC________CUCUCGUUCUUUCCCACA ..........................................................(((((.................................................)))))... ( -4.47 = -4.47 + 0.00)

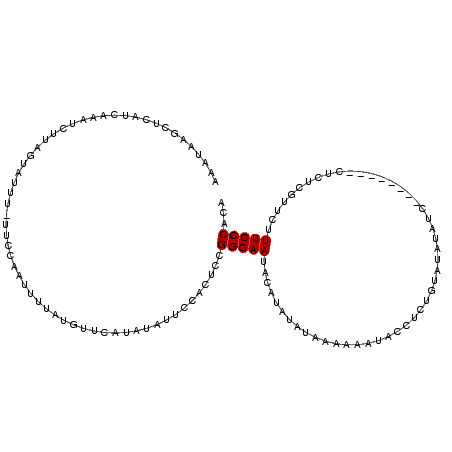
| Location | 574,495 – 574,602 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.84 |
| Mean single sequence MFE | -23.38 |
| Consensus MFE | -9.44 |
| Energy contribution | -9.76 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.40 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.681856 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 574495 107 - 22224390 UGUGGGAAAGAACG------------GGUAUAUACAGACGUAAAUUUUAUAUAUGUAUUCCCGGAGUGGAAUAUAUGAACAUAAAAUUGGAA-AACUGAUAAGAUAUGAUGAGCUUAUUU .......(((..((------------..(((((.(((.....((((((((.((((((((((......))))))))))...))))))))....-..))).....))))).))..))).... ( -17.20) >DroSec_CAF1 38287 111 - 1 UGUGGGAAAGAACGAGAG--------GAUAUAUACAGAGGUAUUUUUUAUAUAUGUAUUCCCGGAGGGGAAUAUAUAUACAUAAAAUUGGAA-AAAUACUAAGAUUUGAUGAGCUUCUUU .............(((((--------(...(((.((((((((((((((.(((((((((((((....)))))))))))))((......)))))-)))))))....)))))))..)))))). ( -29.60) >DroSim_CAF1 37820 111 - 1 UGUGGGAAAGAACGAGAG--------GAUAUAUACAGAGGUAUUUUUUAUAUAUGUAUUCCCGGAGUGGAAUAUAUAUACAUAAAAUUGGGA-AAAUACUAAGAUUUGAUGAGCUUCUUU .............(((((--------(...(((.((((((((((((((.((((((((((((......))))))))))))((......)))))-)))))))....)))))))..)))))). ( -25.30) >DroEre_CAF1 38618 109 - 1 UGUGGGAAGAAGCGAGAG--------G--GUACACAGUGUUAUU-UAUACUUGUAUAUUCCCGGAGUGGAGUAUAUGAAUAUGAAAUUGAAAACAAUACUAAGAUUUGUUGCGCUUAUUU .........(((((...(--------(--((....((((((.((-((((.(..((((((((......))))))))..).))))))..(....).))))))...))))....))))).... ( -22.40) >DroYak_CAF1 41916 102 - 1 UGUGGGAAUGGGCGAGAGAACGAGAGGUUGCAUACAGUGUUAUU-UAU------AUAUUCCCGGAGUGGAGUGUAUGAAAAUGAAAUUGGAA-CCAUACUUGGAU----------UAUUA ...(((.((((((((.....(....).))))...((((.(((((-(((------(((((((......)))))))))))..)))).))))...-)))).)))....----------..... ( -22.40) >consensus UGUGGGAAAGAACGAGAG________GAUAUAUACAGAGGUAUUUUUUAUAUAUGUAUUCCCGGAGUGGAAUAUAUGAACAUAAAAUUGGAA_AAAUACUAAGAUUUGAUGAGCUUAUUU .................................................((((((((((((......))))))))))))......................................... ( -9.44 = -9.76 + 0.32)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:37:36 2006