
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,590,654 – 4,590,762 |
| Length | 108 |
| Max. P | 0.990190 |

| Location | 4,590,654 – 4,590,762 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 75.19 |
| Mean single sequence MFE | -27.50 |
| Consensus MFE | -16.38 |
| Energy contribution | -16.35 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.60 |
| SVM decision value | 2.20 |
| SVM RNA-class probability | 0.990190 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
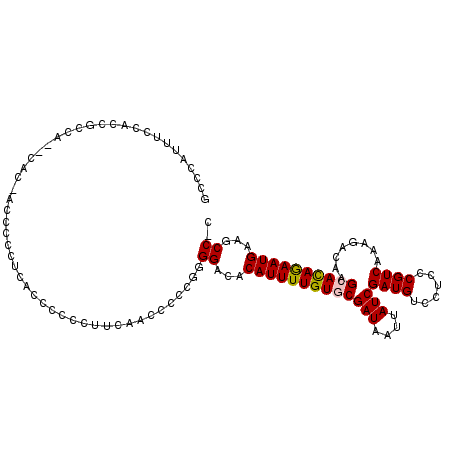
>X_DroMel_CAF1 4590654 108 - 22224390 GCCCAUUUCCACAACCACCCACCAACCC----CCCCCCUCCAA-CCCCGGGGACACAUUUUGUGCGAUAAUUAUCGAUGUCCUGCCGUCAAAGACAAGCAUAGAAUGAAGCC-C ((..........................----..((((.....-....))))...(((((((((((((....)))((((......))))........))))))))))..)).-. ( -22.90) >DroPse_CAF1 18172 106 - 1 GCCCAUUGCGGAUGCCG--UGU-----CGGCAUCGGCCGUCGGCCCUCCAGGACACAUUUUGUACGAUAAUUAUCGAUGUCCUUCCGUCACAGACAAGCACAAAAUGAAACC-U (((.((.((.(((((((--...-----))))))).)).)).))).....(((...((((((((.((((....)))).((((...........))))...))))))))...))-) ( -33.90) >DroSec_CAF1 14605 111 - 1 GCCCAUUUCCACCUACA--ACCCACCCCCUACCCCCCCUUCAACCCCCGGGGACACAUUUUGUGCGAUAAUUAUCGAUGUCCUCCCGUCAAAGACAAGCACAGAAUGAAGCC-C ((...............--...............((((..........))))...(((((((((((((....)))((((......))))........))))))))))..)).-. ( -24.70) >DroSim_CAF1 13922 111 - 1 GCCCAUUUCCACCCCCA--CACCACCCCCUUACCCCCCUUCAACCCCCGGGGACACAUUUUGUGCGAUAAUUAUCGAUGUCCUCCCGUCAAAGACAAGCACAGAAUGAAGCC-C ((...............--...............((((..........))))...(((((((((((((....)))((((......))))........))))))))))..)).-. ( -24.70) >DroEre_CAF1 17655 99 - 1 GCCCAUUUUCCGC----------CCCCCCUUCCGC----CCCAA-CCCGGGGACACAUUUUGUGCGAUAAUUAUCGAUGUCCGCCCGUCAAAGACAAGCACAGAAUGAAGCCCC ...........((----------..........((----(((..-...)))).).(((((((((((((....)))((((......))))........))))))))))..))... ( -26.20) >DroPer_CAF1 19031 106 - 1 GCCCAUUGCGGAUGCCG--UGU-----CGGCAUCGGCCUUCGGCCCUCCAGGACACAUUUUGUACGAUAAUUAUCGAUGUCCUUCCGUCACAGACAAGCACAAAAUGAAACC-U (((.(..((.(((((((--...-----))))))).))..).))).....(((...((((((((.((((....)))).((((...........))))...))))))))...))-) ( -32.60) >consensus GCCCAUUUCCACCGCCA__CAC_ACCCCCUCACCCCCCUUCAACCCCCGGGGACACAUUUUGUGCGAUAAUUAUCGAUGUCCUCCCGUCAAAGACAAGCACAGAAUGAAGCC_C ..................................................((...(((((((((((((....)))((((......))))........))))))))))...)).. (-16.38 = -16.35 + -0.03)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:14:19 2006