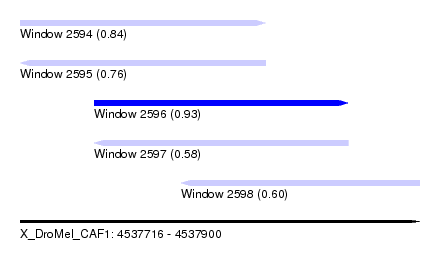
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,537,716 – 4,537,900 |
| Length | 184 |
| Max. P | 0.926379 |

| Location | 4,537,716 – 4,537,829 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.85 |
| Mean single sequence MFE | -28.00 |
| Consensus MFE | -14.68 |
| Energy contribution | -16.44 |
| Covariance contribution | 1.76 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.842455 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4537716 113 + 22224390 CACACUGAAACCCCAA------ACCCACUUUCAUCCCAGUAAACUUGUGGCUUGCACUGCGUUUGGCAAUUUCACAAUUUUGCGGUG-CAGCCUUUGGGUUUUCCGCCCACGCGUCCAUU ......((((.(((((------(...(((........)))........((((.((((((((.(((.........)))...)))))))-))))))))))).))))(((....)))...... ( -31.80) >DroSec_CAF1 706 111 + 1 C--ACUGAAACCCCAA------ACCAACUUUCCUCCCAGUAAACUUGUGGCUUGCACUGCGUUUGGCAAUUUCACAAUUUUGCGGUG-CAGCCUUUGGGUUUUCCGCCCACGCGUCUGUU .--...((((.(((((------(...(((........)))........((((.((((((((.(((.........)))...)))))))-))))))))))).))))(((....)))...... ( -31.80) >DroEre_CAF1 6054 103 + 1 C--A-UGUAACC-------------CACUUUCCUCCCAGUAAACUUGUGGCUUGCACUGCGUUUGGCAAUUUCGCAAUUUUGCGAUG-CAGCCUUUGGGUUUCCCGCCCACAAGUCCAUU .--.-.......-------------.................((((((((((.(((.(((.....)))...(((((....)))))))-)))))...((((.....))))))))))..... ( -27.30) >DroYak_CAF1 6616 110 + 1 C--A-UGUAACCCCAA------ACUCACUUUUCUCCCAGUAAACUUGUGGUUUGCACUGCGUUUGGCAAUUUCACAAUUUUGCGGUG-CAGCCUUUGGGUUUUCCGCCCACGUUUCCAUU .--.-...........------................(.((((....((((.((((((((.(((.........)))...)))))))-)))))..(((((.....))))).)))).)... ( -25.80) >DroAna_CAF1 5727 99 + 1 UAAGAUGCAACACCGACCGACCACCCAC-----UCUCUGUACAAUUGCGG-----------UUUGGCAAUUUCACAAUUUUGCGGUUUUGGUCUUUGGGCU----GCCCAC-UGUCCACU ..............((((((..(((((.-----....(((..((((((..-----------....))))))..)))....)).))).))))))..(((((.----......-.))))).. ( -23.30) >consensus C__A_UGAAACCCCAA______ACCCACUUUCCUCCCAGUAAACUUGUGGCUUGCACUGCGUUUGGCAAUUUCACAAUUUUGCGGUG_CAGCCUUUGGGUUUUCCGCCCACGCGUCCAUU ................................................((((.((((((((.(((.........)))...))))))).)))))..(((((.....))))).......... (-14.68 = -16.44 + 1.76)



| Location | 4,537,716 – 4,537,829 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.85 |
| Mean single sequence MFE | -32.72 |
| Consensus MFE | -11.96 |
| Energy contribution | -13.16 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.37 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.757151 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4537716 113 - 22224390 AAUGGACGCGUGGGCGGAAAACCCAAAGGCUG-CACCGCAAAAUUGUGAAAUUGCCAAACGCAGUGCAAGCCACAAGUUUACUGGGAUGAAAGUGGGU------UUGGGGUUUCAGUGUG ......(((....)))((((.(((...(((((-(((((((....))))....(((.....))))))).)))).((((..((((........))))..)------))))))))))...... ( -35.70) >DroSec_CAF1 706 111 - 1 AACAGACGCGUGGGCGGAAAACCCAAAGGCUG-CACCGCAAAAUUGUGAAAUUGCCAAACGCAGUGCAAGCCACAAGUUUACUGGGAGGAAAGUUGGU------UUGGGGUUUCAGU--G ......(((....)))((((.(((((((((((-(((((((....))))....(((.....))))))).)))).(((.(((.((....))))).))).)------))))).))))...--. ( -34.20) >DroEre_CAF1 6054 103 - 1 AAUGGACUUGUGGGCGGGAAACCCAAAGGCUG-CAUCGCAAAAUUGCGAAAUUGCCAAACGCAGUGCAAGCCACAAGUUUACUGGGAGGAAAGUG-------------GGUUACA-U--G ..(((((((((..(.((....)))...(((((-(.(((((....))))).(((((.....))))))).)))))))))))))...........(((-------------.....))-)--. ( -32.00) >DroYak_CAF1 6616 110 - 1 AAUGGAAACGUGGGCGGAAAACCCAAAGGCUG-CACCGCAAAAUUGUGAAAUUGCCAAACGCAGUGCAAACCACAAGUUUACUGGGAGAAAAGUGAGU------UUGGGGUUACA-U--G .(((....)))........(((((...((.((-(((((((....))))....(((.....))))))))..)).(((..(((((........)))))..------))))))))...-.--. ( -29.40) >DroAna_CAF1 5727 99 - 1 AGUGGACA-GUGGGC----AGCCCAAAGACCAAAACCGCAAAAUUGUGAAAUUGCCAAA-----------CCGCAAUUGUACAGAGA-----GUGGGUGGUCGGUCGGUGUUGCAUCUUA ........-....((----((((((..(((((...((((....(((((.((((((....-----------..)))))).)))))...-----)))).)))))..).)).)))))...... ( -32.30) >consensus AAUGGACACGUGGGCGGAAAACCCAAAGGCUG_CACCGCAAAAUUGUGAAAUUGCCAAACGCAGUGCAAGCCACAAGUUUACUGGGAGGAAAGUGGGU______UUGGGGUUACA_U__G .........((((..((.....))...(((......((((....)))).....)))..............))))...((((((........))))))....................... (-11.96 = -13.16 + 1.20)

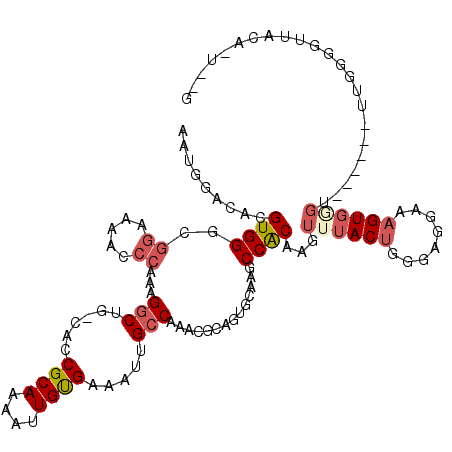

| Location | 4,537,750 – 4,537,867 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 83.76 |
| Mean single sequence MFE | -30.35 |
| Consensus MFE | -25.67 |
| Energy contribution | -25.55 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.926379 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4537750 117 + 22224390 AAACUUGUGGCUUGCACUGCGUUUGGCAAUUUCACAAUUUUGCGGUGCAGCCUUUGGGUUUUCCGCCCACGCGUCCAUUUCCGCAGCCCAUUU--UCACGCCCCGCAUUCAACGCCCAU .....((.(((.(((((((((.(((.........)))...)))))))))((...(((((.....))))).(((........))).........--.........)).......))))). ( -31.30) >DroSec_CAF1 738 117 + 1 AAACUUGUGGCUUGCACUGCGUUUGGCAAUUUCACAAUUUUGCGGUGCAGCCUUUGGGUUUUCCGCCCACGCGUCUGUUUUCACAGCCCAUUU--UCACGCUCCGCAUCCAACGCCCAU .....((.(((.(((((((((.(((.........)))...)))))))))((...(((((.....))))).((((((((....)))).......--..))))...)).......))))). ( -33.20) >DroEre_CAF1 6078 107 + 1 AAACUUGUGGCUUGCACUGCGUUUGGCAAUUUCGCAAUUUUGCGAUGCAGCCUUUGGGUUUCCCGCCCACAAGUCCAUUUCCACAGCC-AUUUCCUCACAGCCCAC-----------AU ..((((((((((.(((.(((.....)))...(((((....))))))))))))...((((.....))))))))))..............-.................-----------.. ( -27.30) >DroYak_CAF1 6647 114 + 1 AAACUUGUGGUUUGCACUGCGUUUGGCAAUUUCACAAUUUUGCGGUGCAGCCUUUGGGUUUUCCGCCCACGUUUCCAUUUCCACAGCC-AUUUCCUUUU-GCCCCUAUGCAA---CCAU ....((((((.((((((((((.(((.........)))...))))))))))....(((((.....)))))...........))))))..-........((-((......))))---.... ( -29.60) >consensus AAACUUGUGGCUUGCACUGCGUUUGGCAAUUUCACAAUUUUGCGGUGCAGCCUUUGGGUUUUCCGCCCACGCGUCCAUUUCCACAGCC_AUUU__UCACGCCCCGCAU_CAA___CCAU ....((((((((.((((((((.(((.........)))...))))))))))))..(((((.....))))).............))))................................. (-25.67 = -25.55 + -0.12)

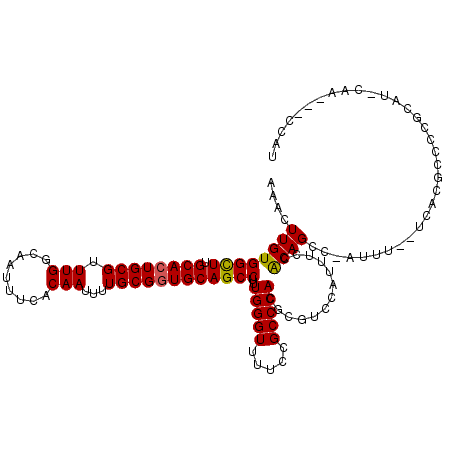
| Location | 4,537,750 – 4,537,867 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 83.76 |
| Mean single sequence MFE | -34.80 |
| Consensus MFE | -21.77 |
| Energy contribution | -21.90 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.576962 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4537750 117 - 22224390 AUGGGCGUUGAAUGCGGGGCGUGA--AAAUGGGCUGCGGAAAUGGACGCGUGGGCGGAAAACCCAAAGGCUGCACCGCAAAAUUGUGAAAUUGCCAAACGCAGUGCAAGCCACAAGUUU .((((((((...(((((..(((..--..)))..)))))..))))..(((....))).....))))..((((((((((((....))))....(((.....))))))).))))........ ( -37.00) >DroSec_CAF1 738 117 - 1 AUGGGCGUUGGAUGCGGAGCGUGA--AAAUGGGCUGUGAAAACAGACGCGUGGGCGGAAAACCCAAAGGCUGCACCGCAAAAUUGUGAAAUUGCCAAACGCAGUGCAAGCCACAAGUUU .(((((......(((((.(((.(.--...((((((((....)))).(((....))).....))))....)))).)))))...((((...(((((.....)))))))))))).))..... ( -37.10) >DroEre_CAF1 6078 107 - 1 AU-----------GUGGGCUGUGAGGAAAU-GGCUGUGGAAAUGGACUUGUGGGCGGGAAACCCAAAGGCUGCAUCGCAAAAUUGCGAAAUUGCCAAACGCAGUGCAAGCCACAAGUUU ..-----------...((((((......))-))))((((...((.(((.(((.(.((....)))...(((....(((((....)))))....)))...)))))).))..))))...... ( -35.10) >DroYak_CAF1 6647 114 - 1 AUGG---UUGCAUAGGGGC-AAAAGGAAAU-GGCUGUGGAAAUGGAAACGUGGGCGGAAAACCCAAAGGCUGCACCGCAAAAUUGUGAAAUUGCCAAACGCAGUGCAAACCACAAGUUU .(((---((((((.(.(((-((.....(((-...(((((..(((....)))..((((....((....)))))).)))))..)))......)))))...)...)))).)))))....... ( -30.00) >consensus AUGG___UUG_AUGCGGGCCGUGA__AAAU_GGCUGUGGAAAUGGACGCGUGGGCGGAAAACCCAAAGGCUGCACCGCAAAAUUGUGAAAUUGCCAAACGCAGUGCAAGCCACAAGUUU .............................((.((((((............((((.......))))..(((.....((((....)))).....)))...)))))).))............ (-21.77 = -21.90 + 0.13)


| Location | 4,537,790 – 4,537,900 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 68.97 |
| Mean single sequence MFE | -29.83 |
| Consensus MFE | -16.39 |
| Energy contribution | -17.07 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.598679 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4537790 110 - 22224390 AA-------UAGAGAGAAAGGAAAACUGCAACGCAGCUGCAUGGGCGUUGAAUGCGGGGCGUGA--AAAUGGGCUGCGGAAAUGGACGCGUGGGCGGAAAACCCAAAGGCUGCACCGCA ..-------..................((...((((((...((((((((...(((((..(((..--..)))..)))))..))))..(((....))).....))))..))))))...)). ( -33.50) >DroSec_CAF1 778 117 - 1 AAAAAAAUACAGAGAGGAAGGAAAACUGCAACGCAGUUGCAUGGGCGUUGGAUGCGGAGCGUGA--AAAUGGGCUGUGAAAACAGACGCGUGGGCGGAAAACCCAAAGGCUGCACCGCA ............................((((((..........))))))..(((((.(((.(.--...((((((((....)))).(((....))).....))))....)))).))))) ( -32.80) >DroEre_CAF1 6118 91 - 1 AA--------AAG---GGGGGCAAAUA-----GCUGUUGGAU-----------GUGGGCUGUGAGGAAAU-GGCUGUGGAAAUGGACUUGUGGGCGGGAAACCCAAAGGCUGCAUCGCA ..--------...---((.(((.....-----))).)).(((-----------(..(.((.((.......-(.(((..(........)..))).)((....)))).)).)..))))... ( -23.20) >consensus AA________AGAGAGGAAGGAAAACUGCAACGCAGUUGCAUGGGCGUUG_AUGCGGGGCGUGA__AAAUGGGCUGUGGAAAUGGACGCGUGGGCGGAAAACCCAAAGGCUGCACCGCA ............................((((((..........))))))...((((.((((......)).(((((((........))).((((.......))))..)))))).)))). (-16.39 = -17.07 + 0.68)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:14:02 2006