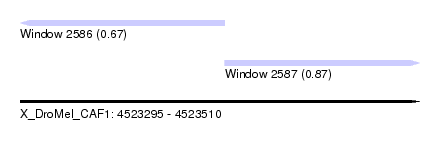
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,523,295 – 4,523,510 |
| Length | 215 |
| Max. P | 0.872592 |

| Location | 4,523,295 – 4,523,405 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 78.62 |
| Mean single sequence MFE | -15.28 |
| Consensus MFE | -1.30 |
| Energy contribution | -2.18 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.09 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.668565 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4523295 110 - 22224390 GCAUCACCACAACAGCCCCAGAAAAUAUCACUUAUUCGAUUUGGCUAUGA--AAGUAUCUUACAAUUCCAACUUUUCAAAGUAACUAGCUUUCAAAGAUCACUUAAAUCUAC (.(((........((((......((((.....))))......)))).(((--((((...((((.................))))...)))))))..))))............ ( -12.73) >DroSec_CAF1 18505 97 - 1 GCAUCACCGCAACAACCCUAGAAAAUAUCACUU-UUUGAUUUGGCUACAAGCAUGUAUCU--------------UUCAAUGUAACUAGCUUUUAAAGAUCACUCAAAUCUAC ((......))........((((.........((-(((((...(((((...((((......--------------....))))...))))).))))))).........)))). ( -11.97) >DroSim_CAF1 10939 110 - 1 GCAUCACCGCAACAACCCCAUAAAAUAUCA-UU-UUUAAUUUGGCUACGAGCAUGUAUCUUACAAUUCCAACUUUUCAAAGUAACUAGCUUUCAAAGAUCAAUCAAACCUAC ((......))..........(((((.....-.)-)))).(((((....((((.((((...))))......((((....)))).....)))))))))................ ( -7.40) >DroEre_CAF1 10021 110 - 1 GCAUCACCUCAACAACCCCAGAAAAUAUCACUUUUUGGAUGUGUUUACGA--AAGUAUCUUACAAUUGCAAUUCUUCAAAGCAACUAACUUCCGAAGAUCAAUUAAAUCUAC (((..............(((((((.......))))))).((((..(((..--..)))...))))..)))...(((((.(((.......)))..))))).............. ( -18.50) >DroYak_CAF1 18783 110 - 1 GCAUCAGCGCAGCAACCCCAGAAAAUAUAAUUUUCUGGUUUUGCUUACGA--AAGUAUUUUACAAUUCCCACUCCUCAAAGCAACUAGCUUUCGCAGAUCAAUCAAAUCCAC (.(((.(((.(((((..(((((((((...)))))))))..)))))(((..--..)))....................(((((.....)))))))).))))............ ( -25.80) >consensus GCAUCACCGCAACAACCCCAGAAAAUAUCACUU_UUGGAUUUGGCUACGA__AAGUAUCUUACAAUUCCAACUCUUCAAAGUAACUAGCUUUCAAAGAUCAAUCAAAUCUAC .....................................((((((..(((......)))....................(((((.....)))))...........))))))... ( -1.30 = -2.18 + 0.88)

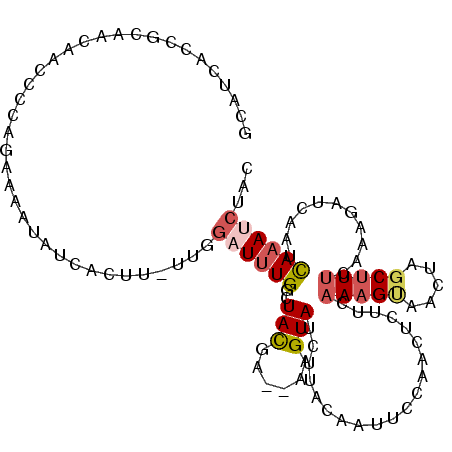

| Location | 4,523,405 – 4,523,510 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.07 |
| Mean single sequence MFE | -26.46 |
| Consensus MFE | -11.15 |
| Energy contribution | -13.77 |
| Covariance contribution | 2.61 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.42 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.872592 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4523405 105 + 22224390 CACCCCCU-UUUCCAG-G-------CAACUGAACCCGUUCGCCCCCUCU----CUGUCGCACCCACACAGUGCAUUCGAUGGAAGGUGACACGUCACUUUCCACGACUCCUA--CCCUCU ........-....(((-.-------...)))..................----..(((((((.......))))....(.((((((((((....)))))))))))))).....--...... ( -27.30) >DroSec_CAF1 18602 97 + 1 CACCCCCU-UUUCCAGGG-------CAACUGAACCCGUUCGCCCCCUCU----CUGUCGCACCCACACAGUGCAU---------GGUGACACGUCACUUUCCACGACUCCUA--CCCUCU ........-......(((-------((((.......))).)))).....----..(((((((.......))))..---------(((((....)))))......))).....--...... ( -20.40) >DroSim_CAF1 11049 106 + 1 CACCCCCU-UUUCCAGGG-------CAACUGAACCCGUUCGCCCCCUCU----CUGUCGCACCCACACAGUGCAUUUGAUGGAAGGUGACACGUCACUUUCCACGACUCCUA--CCCUCU ........-......(((-------((((.......))).)))).....----..(((((((.......))))......((((((((((....)))))))))).))).....--...... ( -30.20) >DroEre_CAF1 10131 107 + 1 CACCCCCUUUUUCCAGGG-------CAACUGAACCCGUUCGCCCGCUCU----CUGUCGCACCCACACAAUGCAUUUGAUGGAAGGUGACACGUCACUUUCCACGGCCCCUA--CCCUCU ...............(((-------((((.......))).)))).....----..((((........(((.....))).((((((((((....)))))))))))))).....--...... ( -26.70) >DroYak_CAF1 18893 113 + 1 CACCCCGU-UUUCCAGGGGGUGAAUUAACUGAACCCGUUCGCCCCCUCU----CUGUCGCACCCACACAAUGCAUCUGCUGGAAGGUGACACGUCACUUUCCACGACCCCUA--CCCUCU ........-.....(((((((((((...........)))))))))))..----..((((............((....))((((((((((....)))))))))))))).....--...... ( -36.30) >DroAna_CAF1 23711 112 + 1 CACCCCUU-UGUUUAGAG------CCACCCCAACUCCUACUCUCCUACUUCUACUCUCUCGCUCGCACACUGCAU-UGAUAAAUGGUGACACGUAGGUCCCCCUGACCCGCAUAUCCCCA ((((..((-((((..(((------(...................................))))((.....))..-.)))))).))))....(..((((.....))))..)......... ( -17.85) >consensus CACCCCCU_UUUCCAGGG_______CAACUGAACCCGUUCGCCCCCUCU____CUGUCGCACCCACACAGUGCAUUUGAUGGAAGGUGACACGUCACUUUCCACGACCCCUA__CCCUCU ..............((((............(((....)))...............((((((.........)))......((((((((((....)))))))))).))).......)))).. (-11.15 = -13.77 + 2.61)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:13:52 2006