
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,482,992 – 4,483,087 |
| Length | 95 |
| Max. P | 0.997041 |

| Location | 4,482,992 – 4,483,087 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 83.13 |
| Mean single sequence MFE | -30.48 |
| Consensus MFE | -20.22 |
| Energy contribution | -20.22 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.743649 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
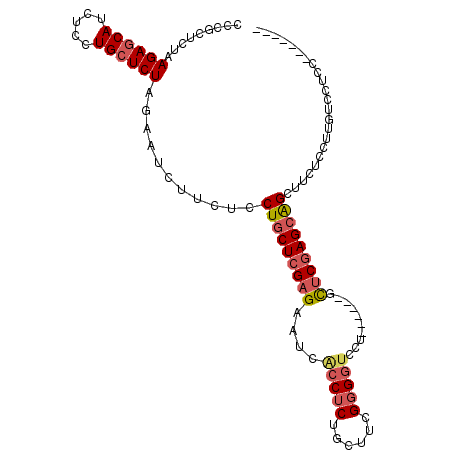
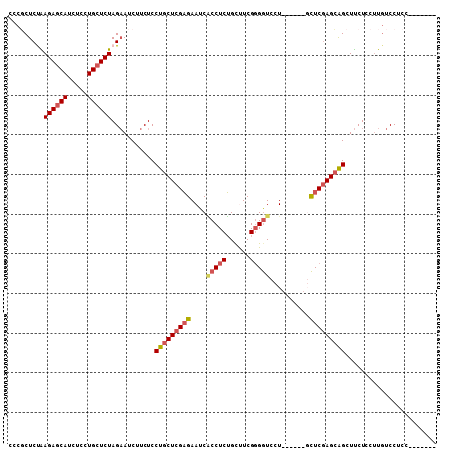
>X_DroMel_CAF1 4482992 95 + 22224390 -------AGAGGACACGGAGAAGCUGCUCGAGC------AGGACCGCGAAGCAGAGGUGAUUCUCGAGCAGGAGAAGAUUCUAGAGCAGGAGAUGGUCUCAGAGCGGG -------........((......(((((((((.------(..(((.(......).)))..).))))))))).......(((((((.((.....)).))).)))))).. ( -28.00) >DroSec_CAF1 1283 101 + 1 -------GGAGGACAAGGAGAAGCUGCUCGAGCUGCUCGAGGACCCCGAAACAGAGGUGAUUCUCGAGCAGGAGAAGAUUCUAGAGCAGGAGAUGCUCUUGGAGCGGG -------................((((((...(((((((((((.(((........)).).))))))))))).........(.((((((.....)))))).))))))). ( -39.50) >DroSim_CAF1 315 95 + 1 -------GGAGGACAAGGAGAAGCUGCUCGAGC------AGGACCCCGAAACAGAGGUGAUUCUCGAGCAGGAGAAGAUUCUAGAGCAGGAGAUGCUCUUGGAGCGGG -------................(((((((((.------(..(((.(......).)))..).))))))))).....(.((((((((((.....))))).))))))... ( -31.00) >DroEre_CAF1 7581 95 + 1 -------GGAGGAGAAGGAGAAGCCUCUUGAGA------AGGACCCCGAGGCAGAGGCGAUUCUCGAGCAGGAGAGGAUUCUAGAGCAGGAGAUGCUCUUAGAGAGGG -------................((((((((((------(...((.(......).))...)))))))).)))......((((((((((.....))))).))))).... ( -26.90) >DroYak_CAF1 5903 102 + 1 AGGUGGAGGAGGACAAGGAGGAACUGCUCGAAA------AAGAUCCCGAGGCCGAGGAGAUUCGCGAGCAGGAGAGGAUGCUAGAGCAAGAGAUGCUCUCAGAGAGGG .......................(((((((...------((..(((((....)).)))..))..)))))))(((((.((.((........)))).)))))........ ( -27.00) >consensus _______GGAGGACAAGGAGAAGCUGCUCGAGC______AGGACCCCGAAGCAGAGGUGAUUCUCGAGCAGGAGAAGAUUCUAGAGCAGGAGAUGCUCUUAGAGCGGG .......................((((((.........................(((...(((((......)))))...)))((((((.....))))))..)))))). (-20.22 = -20.22 + 0.00)



| Location | 4,482,992 – 4,483,087 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 83.13 |
| Mean single sequence MFE | -28.11 |
| Consensus MFE | -19.60 |
| Energy contribution | -20.56 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.58 |
| Structure conservation index | 0.70 |
| SVM decision value | 2.79 |
| SVM RNA-class probability | 0.997041 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4482992 95 - 22224390 CCCGCUCUGAGACCAUCUCCUGCUCUAGAAUCUUCUCCUGCUCGAGAAUCACCUCUGCUUCGCGGUCCU------GCUCGAGCAGCUUCUCCGUGUCCUCU------- ...((...(((.....)))..))..............(((((((((....(((.(......).)))...------.)))))))))................------- ( -23.60) >DroSec_CAF1 1283 101 - 1 CCCGCUCCAAGAGCAUCUCCUGCUCUAGAAUCUUCUCCUGCUCGAGAAUCACCUCUGUUUCGGGGUCCUCGAGCAGCUCGAGCAGCUUCUCCUUGUCCUCC------- ...((((..((((((.....))))))...........(((((((((....(((((......))))).)))))))))...))))..................------- ( -34.50) >DroSim_CAF1 315 95 - 1 CCCGCUCCAAGAGCAUCUCCUGCUCUAGAAUCUUCUCCUGCUCGAGAAUCACCUCUGUUUCGGGGUCCU------GCUCGAGCAGCUUCUCCUUGUCCUCC------- .........((((((.....))))))...........(((((((((....(((((......)))))...------.)))))))))................------- ( -30.20) >DroEre_CAF1 7581 95 - 1 CCCUCUCUAAGAGCAUCUCCUGCUCUAGAAUCCUCUCCUGCUCGAGAAUCGCCUCUGCCUCGGGGUCCU------UCUCAAGAGGCUUCUCCUUCUCCUCC------- .((((((((.(((((.....)))))))))..............(((((..(((((......)))))..)------))))..))))................------- ( -27.20) >DroYak_CAF1 5903 102 - 1 CCCUCUCUGAGAGCAUCUCUUGCUCUAGCAUCCUCUCCUGCUCGCGAAUCUCCUCGGCCUCGGGAUCUU------UUUCGAGCAGUUCCUCCUUGUCCUCCUCCACCU .........((((((.....)))))).(((.......(((((((.(((..(((.((....)))))...)------)).)))))))........)))............ ( -25.06) >consensus CCCGCUCUAAGAGCAUCUCCUGCUCUAGAAUCUUCUCCUGCUCGAGAAUCACCUCUGCUUCGGGGUCCU______GCUCGAGCAGCUUCUCCUUGUCCUCC_______ .........((((((.....))))))...........(((((((((....(((((......)))))..........)))))))))....................... (-19.60 = -20.56 + 0.96)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:13:48 2006