
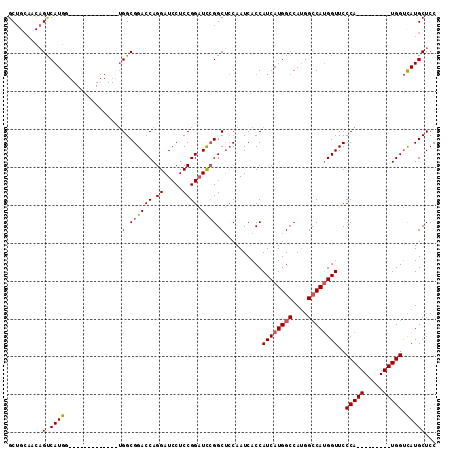
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,468,869 – 4,469,006 |
| Length | 137 |
| Max. P | 0.964808 |

| Location | 4,468,869 – 4,468,970 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 73.27 |
| Mean single sequence MFE | -39.33 |
| Consensus MFE | -21.37 |
| Energy contribution | -22.27 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.911292 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4468869 101 + 22224390 GCUGCAACAGUCAUGG------------CUGGCGGACCAGGAUCCUCCGGAUUCGGCUCCAAUCACCAUCAUGGCCAUGGCCAUGGUUCCCAUGGUUCCCAUGGUCAUGCUCC ...((..((((....)------------)))))(((((.((((((...)))))))).))).........(((((((((((((((((...))))))...))))))))))).... ( -41.70) >DroSec_CAF1 70507 104 + 1 GCUGCAACAGUCAUGGGUCAGGUGCCGGUUGGCGGACCAGGAUCCUCCGGAUCGGGCUCCAAUCACCAUCAUGGCCACGGCCAUGGUUCCCA---------UGGUUAUGCUCC ...(((..(..((((((...((((((....)))(((((..(((((...))))).)).)))....)))((((((((....)))))))).))))---------))..).)))... ( -44.50) >DroYak_CAF1 71387 83 + 1 GC------CGCCAUG---------------GGCGGACCAGGAUCCUCCGGUUCCGGCAGCAAUCACCAUCAUGGCCAUGGGCACGGUUCCCA---------UGGACAUGCUCC ((------((((...---------------)))(((((.((.....))))))).)))............((((.(((((((.......))))---------))).)))).... ( -31.80) >consensus GCUGCAACAGUCAUGG_____________UGGCGGACCAGGAUCCUCCGGAUCCGGCUCCAAUCACCAUCAUGGCCAUGGCCAUGGUUCCCA_________UGGUCAUGCUCC .........(.((((...............(.((((((.((.....)))).)))).)..........((((((((....))))))))..(((((.....))))).)))))... (-21.37 = -22.27 + 0.89)


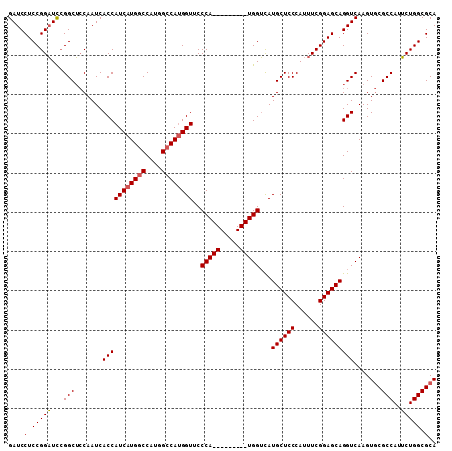
| Location | 4,468,897 – 4,469,006 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 86.79 |
| Mean single sequence MFE | -43.20 |
| Consensus MFE | -33.98 |
| Energy contribution | -34.54 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.54 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.964808 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4468897 109 + 22224390 GAUCCUCCGGAUUCGGCUCCAAUCACCAUCAUGGCCAUGGCCAUGGUUCCCAUGGUUCCCAUGGUCAUGCUCCCAUUUCGGAGCAGGUCAAGUGCGCCAUUCUGGCGCA (((((((((((...((.........))..(((((((((((((((((...))))))...))))))))))).......)))))))..))))...((((((.....)))))) ( -48.10) >DroSec_CAF1 70547 100 + 1 GAUCCUCCGGAUCGGGCUCCAAUCACCAUCAUGGCCACGGCCAUGGUUCCCA---------UGGUUAUGCUCCCAUUUCGGAGCAGGUCAAGUGCGCCAUUCUGGCGCA (((((...)))))((..........))....(((((..((((((((...)))---------))))).((((((......)))))))))))..((((((.....)))))) ( -43.30) >DroYak_CAF1 71406 100 + 1 GAUCCUCCGGUUCCGGCAGCAAUCACCAUCAUGGCCAUGGGCACGGUUCCCA---------UGGACAUGCUCCCAUUUCGGAGCAGGUCAAGUACGCCAUACUGGCGCA (((.(((((....))).))..))).(((..(((((((((((.......))))---------))(((.((((((......)))))).)))......)))))..))).... ( -38.20) >consensus GAUCCUCCGGAUCCGGCUCCAAUCACCAUCAUGGCCAUGGCCAUGGUUCCCA_________UGGUCAUGCUCCCAUUUCGGAGCAGGUCAAGUGCGCCAUUCUGGCGCA ....(.(((((...(((.......(((((((((((....))))))))..(((((.....)))))...((((((......))))))))).......)))..))))).).. (-33.98 = -34.54 + 0.56)


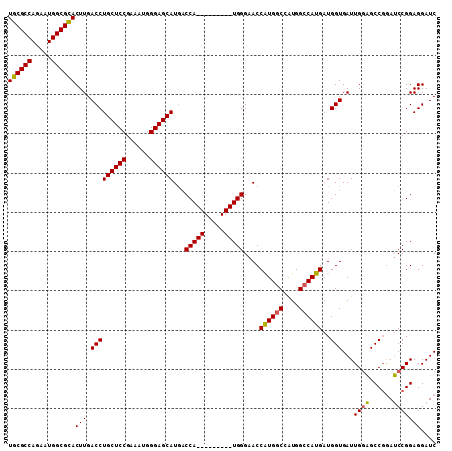
| Location | 4,468,897 – 4,469,006 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 86.79 |
| Mean single sequence MFE | -44.43 |
| Consensus MFE | -36.55 |
| Energy contribution | -36.67 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932181 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4468897 109 - 22224390 UGCGCCAGAAUGGCGCACUUGACCUGCUCCGAAAUGGGAGCAUGACCAUGGGAACCAUGGGAACCAUGGCCAUGGCCAUGAUGGUGAUUGGAGCCGAAUCCGGAGGAUC ((((((.....)))))).....((((((((((.((.(...((((.((((((...((((((...)))))))))))).)))).).))..)))))))((....)).)))... ( -46.30) >DroSec_CAF1 70547 100 - 1 UGCGCCAGAAUGGCGCACUUGACCUGCUCCGAAAUGGGAGCAUAACCA---------UGGGAACCAUGGCCGUGGCCAUGAUGGUGAUUGGAGCCCGAUCCGGAGGAUC ((((((.....)))))).....((((((((......)))))...((((---------((....)((((((....)))))))))))((((((...))))))...)))... ( -42.10) >DroYak_CAF1 71406 100 - 1 UGCGCCAGUAUGGCGUACUUGACCUGCUCCGAAAUGGGAGCAUGUCCA---------UGGGAACCGUGCCCAUGGCCAUGAUGGUGAUUGCUGCCGGAACCGGAGGAUC ..(((((.((((((......(((.((((((......)))))).)))((---------((((.......)))))))))))).)))))....((.(((....))).))... ( -44.90) >consensus UGCGCCAGAAUGGCGCACUUGACCUGCUCCGAAAUGGGAGCAUGACCA_________UGGGAACCAUGGCCAUGGCCAUGAUGGUGAUUGGAGCCGGAUCCGGAGGAUC ((((((.....))))))(((.(((((((((......))))))...(((((.....)))))....((((((....))))))..)))...((((......))))))).... (-36.55 = -36.67 + 0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:13:35 2006