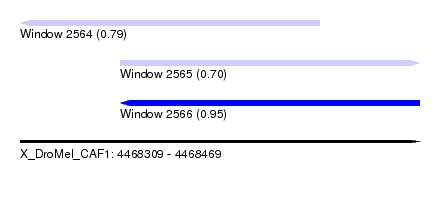
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,468,309 – 4,468,469 |
| Length | 160 |
| Max. P | 0.954464 |

| Location | 4,468,309 – 4,468,429 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.06 |
| Mean single sequence MFE | -58.97 |
| Consensus MFE | -54.38 |
| Energy contribution | -54.17 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.787449 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4468309 120 - 22224390 CGCCAGCACCAAAGAGUUGCUGCUGGGCCAGUAGCAGCUGUUGCUGCUCAAUGGCGGCGGCCAGGUCAGGAGUUCCAACGCCGCCAGGCGUCACUGGAGCAACUGCAGCGGGCGUCGGUG ..((((((.(((....))).))))))((((..((((((....))))))...))))((((.((..(.(((..(((((((((((....)))))...))))))..))))...)).)))).... ( -59.80) >DroSec_CAF1 69950 120 - 1 CGCCAGCUCCAAAGAGUUGCUGCUGGGCCAGCAGCAGCUGUUGCUGCUCAAUGGCGGCGGCCAGGUCAGGAGUUCCAACGCCGCCAGGCGCCACUGGAGCUACUGCAGCGGGCGUCGGUG (((((((..(((..(((((((((((...))))))))))).)))..)))...((((((((((...))).((....))...)))))))((((((.(((.((...)).)))..)))))))))) ( -59.70) >DroYak_CAF1 70827 120 - 1 CGCCAGCGCCAAAGAGUUGCUGCUGCGCGAGGAGCAACUGUUGCUGCUCAAUGGCGGCGGCCAAGUCAGGUGUUCCAACGCCGCCGGGCGCCACUGGAACCACUGCAGCGGGCGUCGGUG ((((.(((((.((.(((((((.((.....)).))))))).))(((((....((((....)))).....(((((((((.((((....))))....))))).))))))))).))))).)))) ( -57.40) >consensus CGCCAGCACCAAAGAGUUGCUGCUGGGCCAGCAGCAGCUGUUGCUGCUCAAUGGCGGCGGCCAGGUCAGGAGUUCCAACGCCGCCAGGCGCCACUGGAGCAACUGCAGCGGGCGUCGGUG ((((((((.(((..((((((((((.....)))))))))).))).))))...((((((((((...))).((....))...)))))))((((((.(((.(.....).)))..)))))))))) (-54.38 = -54.17 + -0.21)


| Location | 4,468,349 – 4,468,469 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.11 |
| Mean single sequence MFE | -46.07 |
| Consensus MFE | -38.23 |
| Energy contribution | -38.57 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.698213 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4468349 120 + 22224390 CGUUGGAACUCCUGACCUGGCCGCCGCCAUUGAGCAGCAACAGCUGCUACUGGCCCAGCAGCAACUCUUUGGUGCUGGCGGCGAGUAUAUGCAGCAGCUAUUCCGCAGUCUCAUGUUCCA ...((((((....(((((.((((((((((...((((((....))))))..))))..((((.(((....))).)))))))))).))...........((......)).)))....)))))) ( -51.90) >DroSec_CAF1 69990 120 + 1 CGUUGGAACUCCUGACCUGGCCGCCGCCAUUGAGCAGCAACAGCUGCUGCUGGCCCAGCAGCAACUCUUUGGAGCUGGCGGCGAAUACAUGCAGCAGAUAUUCCGCAGUCUCAUGUUUCA ...((((((....(((.((((((((.(((..(((.(((....)))(((((((...)))))))..)))..)))....))))))(((((..((...))..)))))..)))))....)))))) ( -45.10) >DroYak_CAF1 70867 120 + 1 CGUUGGAACACCUGACUUGGCCGCCGCCAUUGAGCAGCAACAGUUGCUCCUCGCGCAGCAGCAACUCUUUGGCGCUGGCGGAGAGUACAUGCAACAGAUAUUCCGCAGCCUUAUGUUCCA ...(((((((.....(((.((((.(((((..(((((((....)))))))...((......)).......))))).)))).)))......(((............)))......))))))) ( -41.20) >consensus CGUUGGAACUCCUGACCUGGCCGCCGCCAUUGAGCAGCAACAGCUGCUACUGGCCCAGCAGCAACUCUUUGGAGCUGGCGGCGAGUACAUGCAGCAGAUAUUCCGCAGUCUCAUGUUCCA ...((((((....(((.((((((((((((...((((((....))))))..))))..(((..(((....)))..)))))))))(((((..((...))..)))))..)))))....)))))) (-38.23 = -38.57 + 0.34)



| Location | 4,468,349 – 4,468,469 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.11 |
| Mean single sequence MFE | -51.10 |
| Consensus MFE | -46.13 |
| Energy contribution | -46.37 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.954464 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4468349 120 - 22224390 UGGAACAUGAGACUGCGGAAUAGCUGCUGCAUAUACUCGCCGCCAGCACCAAAGAGUUGCUGCUGGGCCAGUAGCAGCUGUUGCUGCUCAAUGGCGGCGGCCAGGUCAGGAGUUCCAACG ((((((....((((..((....((....))......((((((((((((.(((..(((((((((((...))))))))))).))).)))....))))))))))).))))....))))))... ( -53.60) >DroSec_CAF1 69990 120 - 1 UGAAACAUGAGACUGCGGAAUAUCUGCUGCAUGUAUUCGCCGCCAGCUCCAAAGAGUUGCUGCUGGGCCAGCAGCAGCUGUUGCUGCUCAAUGGCGGCGGCCAGGUCAGGAGUUCCAACG .((((((((.....((((.....))))..))))).)))(((((((((((....)))))(((..(((((.((((((....)))))))))))..))))))))).......((....)).... ( -51.00) >DroYak_CAF1 70867 120 - 1 UGGAACAUAAGGCUGCGGAAUAUCUGUUGCAUGUACUCUCCGCCAGCGCCAAAGAGUUGCUGCUGCGCGAGGAGCAACUGUUGCUGCUCAAUGGCGGCGGCCAAGUCAGGUGUUCCAACG ((((((((..((((((((((((((....).))))....)))))..(.(((.((.(((((((.((.....)).))))))).))((((((....)))))))))).))))..))))))))... ( -48.70) >consensus UGGAACAUGAGACUGCGGAAUAUCUGCUGCAUGUACUCGCCGCCAGCACCAAAGAGUUGCUGCUGGGCCAGCAGCAGCUGUUGCUGCUCAAUGGCGGCGGCCAGGUCAGGAGUUCCAACG ((((((....((((((((.....)))).........((((((((((((.(((..((((((((((.....)))))))))).))).)))....)))))))))...))))....))))))... (-46.13 = -46.37 + 0.23)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:13:33 2006