
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,465,255 – 4,465,517 |
| Length | 262 |
| Max. P | 0.984353 |

| Location | 4,465,255 – 4,465,375 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.03 |
| Mean single sequence MFE | -52.60 |
| Consensus MFE | -45.17 |
| Energy contribution | -45.55 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.12 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.972259 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4465255 120 + 22224390 ACAACAGCACAGUGGAAUCGGAAAUGGAGCGCCAGCGACGGGACUCCACCACAUCGGAGUCCUCGCUGGAGCAUCUGGAUCUGGGACGUACGCCCAAAAAACUGGGCGGUAACAGUGGCU .....(((.((.(((..(((((..((((((.(((((((..(((((((........)))))))))))))).)).))))..)))))..)...((((((......))))))....)).))))) ( -54.70) >DroSec_CAF1 66906 120 + 1 ACAACAGCACAGUGGAAUCGGAAAUGGAGCGCCAGCGACGUGACUCCACCACAUCGGAGUCCUCGCUGGAGCAUCUGGAUCUGGGCCGUACGCCCAAAAAACUGGGCGGCAACAGUGGCU .....(((.((.(((..(((((..((((((.(((((((.(.((((((........)))))))))))))).)).))))..)))))..)((.((((((......))))))))..)).))))) ( -53.20) >DroEre_CAF1 70648 120 + 1 ACAACAGCACAGCGGAAUCCGAAAUGGAGCGCCAGCGGCGGGACUCCACCACAUCGGAGUCCUCCCUGGAGCAUCUGGAUCUGGGACGUACGCCCAAGAAACUGGGCGGCAACAGUGGCU .......(((.(((...(((((..((((((.((((.((.((((((((........)))))))))))))).)).))))..)).))).))).((((((......))))))......)))... ( -49.60) >DroYak_CAF1 67800 120 + 1 ACAACAGCACAGCGGAGUCGGAAAUGGAGCGUCAGCGGCGGGACUCGACCACAUCGGAGUCCUCGCUGGAGCACCUGGAUCUGGGCCGCACGCCCAAGAAACUGGGCGGCAACAGUGGCU .......(((.((((..(((((...((.((..(((((..(((((((((.....)).))))))))))))..)).))....))))).)))).((((((......))))))......)))... ( -52.90) >consensus ACAACAGCACAGCGGAAUCGGAAAUGGAGCGCCAGCGACGGGACUCCACCACAUCGGAGUCCUCGCUGGAGCAUCUGGAUCUGGGACGUACGCCCAAAAAACUGGGCGGCAACAGUGGCU .......(((.(((...(((((..((((((.(((((((..(((((((........)))))))))))))).)).))))..)))))..))).((((((......))))))......)))... (-45.17 = -45.55 + 0.38)

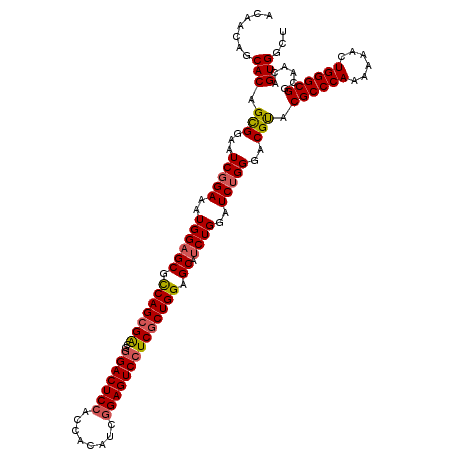
| Location | 4,465,295 – 4,465,397 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 94.12 |
| Mean single sequence MFE | -39.90 |
| Consensus MFE | -36.39 |
| Energy contribution | -36.32 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.984353 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4465295 102 + 22224390 GGACUCCACCACAUCGGAGUCCUCGCUGGAGCAUCUGGAUCUGGGACGUACGCCCAAAAAACUGGGCGGUAACAGUGGCUCUACACAGACCACAUCCACGCC (((((((........)))))))..((((((.....(((.((((....((.((((((......))))))...)).(((....))).)))))))..)))).)). ( -39.10) >DroSec_CAF1 66946 102 + 1 UGACUCCACCACAUCGGAGUCCUCGCUGGAGCAUCUGGAUCUGGGCCGUACGCCCAAAAAACUGGGCGGCAACAGUGGCUCUACACAGACCACAUCCACGCC .((((((........))))))...((((((...((((.....((((((...(((((......)))))(....)..))))))....)))).....)))).)). ( -39.50) >DroEre_CAF1 70688 102 + 1 GGACUCCACCACAUCGGAGUCCUCCCUGGAGCAUCUGGAUCUGGGACGUACGCCCAAGAAACUGGGCGGCAACAGUGGCUCCGCGCAGACCACAUCCACGCC (((((((........)))))))....((((.....(((.((((....((.((((((......))))))))....(((....))).)))))))..)))).... ( -38.90) >DroYak_CAF1 67840 102 + 1 GGACUCGACCACAUCGGAGUCCUCGCUGGAGCACCUGGAUCUGGGCCGCACGCCCAAGAAACUGGGCGGCAACAGUGGCUCCACACAGACCACAUCCACGCC ((((((((.....)).))))))..((((((.....(((.((((((((((..(((((......)))))(....).)))))).....)))))))..)))).)). ( -42.10) >consensus GGACUCCACCACAUCGGAGUCCUCGCUGGAGCAUCUGGAUCUGGGACGUACGCCCAAAAAACUGGGCGGCAACAGUGGCUCCACACAGACCACAUCCACGCC (((((((........)))))))..((((((.....(((.((((....((.((((((......))))))))....(((....))).)))))))..)))).)). (-36.39 = -36.32 + -0.06)



| Location | 4,465,335 – 4,465,437 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 91.01 |
| Mean single sequence MFE | -39.35 |
| Consensus MFE | -33.74 |
| Energy contribution | -36.30 |
| Covariance contribution | 2.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.937142 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4465335 102 + 22224390 CUGGGACGUACGCCCAAAAAACUGGGCGGUAACAGUGGCUCUACACAGACCACAUCCACGCCACAUGAGCUGGCUACGGUGACAUCUUCGCGCAAGCGCAAA ...(((.((.((((((......))))))...)).((((.(((....))))))).)))..((((.......))))...(....).....(((....))).... ( -34.00) >DroSec_CAF1 66986 102 + 1 CUGGGCCGUACGCCCAAAAAACUGGGCGGCAACAGUGGCUCUACACAGACCACAUCCACGCCACAUGAGCUGACUACGGUGACGUCGUCGCGCAAGCGUAAG .(((((.((.((((((......))))))))....((((.(((....)))))))......))).))......(((.(((....))).)))((....))..... ( -38.50) >DroEre_CAF1 70728 102 + 1 CUGGGACGUACGCCCAAGAAACUGGGCGGCAACAGUGGCUCCGCGCAGACCACAUCCACGCCACAUGAGUCGACGACGGUGGCGUCGUCGCGCAAGCGUAAA ...(((.((.((((((......))))))))....((((((......)).)))).)))(((((((....(((...))).)))))))...(((....))).... ( -40.20) >DroYak_CAF1 67880 102 + 1 CUGGGCCGCACGCCCAAGAAACUGGGCGGCAACAGUGGCUCCACACAGACCACAUCCACGCCACAUGAGGUGACGACGGUGGCGUCGUCGCGCAAGCGUAAA .((((((((..(((((......)))))(....).))))).)))..............(((((((....(....)....)))))))...(((....))).... ( -44.70) >consensus CUGGGACGUACGCCCAAAAAACUGGGCGGCAACAGUGGCUCCACACAGACCACAUCCACGCCACAUGAGCUGACGACGGUGACGUCGUCGCGCAAGCGUAAA .((((.(((.((((((......))))))......((((.((......))))))....))))).))......(((((((....)))))))((....))..... (-33.74 = -36.30 + 2.56)



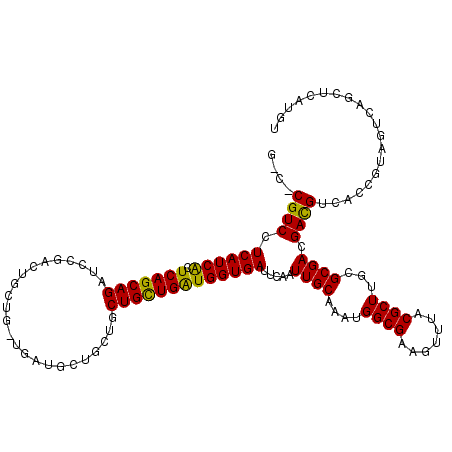
| Location | 4,465,397 – 4,465,517 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.83 |
| Mean single sequence MFE | -40.93 |
| Consensus MFE | -25.21 |
| Energy contribution | -25.43 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.639858 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4465397 120 + 22224390 ACAUGAGCUGGCUACGGUGACAUCUUCGCGCAAGCGCAAAAUUCGCCAUUUGCAAUUGAAUCACCAUCAGCAGCAGCAGCAUCACCAGCAGUCGGAUCUGCUGAGUGAUGAAGACGUGGC .((((.(((((((..(((((....(((((....))(((((........)))))....))))))))...)))..))))..((((((((((((......)))))).))))))....)))).. ( -46.30) >DroSec_CAF1 67048 116 + 1 ACAUGAGCUGACUACGGUGACGUCGUCGCGCAAGCGUAAGCUUCGCCAUUUGCAAUUGAAUCACCAUCAGCAGCAGCAGCAUCAUCAGCAGUCGGAUCUGCUGAGUGAUGAGGACG---- ....((((((((.(((....))).)))((....))...)))))((((..((((..((((.......))))..))))...((((((((((((......))))))).))))).)).))---- ( -42.50) >DroYak_CAF1 67942 108 + 1 ACAUGAGGUGACGACGGUGGCGUCGUCGCGCAAGCGUAAACUUCGCCACUUGCAACUGAAUCACCACCAACAGCAG------------CACUCGGAACUGCUGAGCGAUGAGGACGAGUC ....((((((((((((....)))))))((....))....)))))(((.(((((...((......))....((((((------------.........)))))).)))).).)).)..... ( -34.00) >consensus ACAUGAGCUGACUACGGUGACGUCGUCGCGCAAGCGUAAACUUCGCCAUUUGCAAUUGAAUCACCAUCAGCAGCAGCAGCAUCA_CAGCAGUCGGAUCUGCUGAGUGAUGAGGACG_G_C ...............(((((.((...(((....)))...)).))))).............((..((((..((((((.....................))))))...))))..))...... (-25.21 = -25.43 + 0.23)

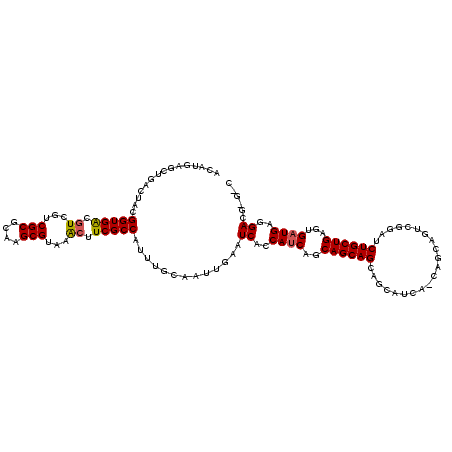

| Location | 4,465,397 – 4,465,517 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.83 |
| Mean single sequence MFE | -43.17 |
| Consensus MFE | -24.81 |
| Energy contribution | -23.93 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.57 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.511568 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4465397 120 - 22224390 GCCACGUCUUCAUCACUCAGCAGAUCCGACUGCUGGUGAUGCUGCUGCUGCUGAUGGUGAUUCAAUUGCAAAUGGCGAAUUUUGCGCUUGCGCGAAGAUGUCACCGUAGCCAGCUCAUGU ...((((...((((((.((((((......))))))))))))..((((..(((.((((((((((..((((.....))))..((((((....)))))))).)))))))))))))))..)))) ( -46.40) >DroSec_CAF1 67048 116 - 1 ----CGUCCUCAUCACUCAGCAGAUCCGACUGCUGAUGAUGCUGCUGCUGCUGAUGGUGAUUCAAUUGCAAAUGGCGAAGCUUACGCUUGCGCGACGACGUCACCGUAGUCAGCUCAUGU ----.((...(((((.(((((((......))))))))))))..)).((((((.((((((((....((((....((((.......))))...))))....)))))))))).))))...... ( -40.90) >DroYak_CAF1 67942 108 - 1 GACUCGUCCUCAUCGCUCAGCAGUUCCGAGUG------------CUGCUGUUGGUGGUGAUUCAGUUGCAAGUGGCGAAGUUUACGCUUGCGCGACGACGCCACCGUCGUCACCUCAUGU ((((.(...(((((((((((((((.......)------------))))))..))))))))..)))))(((((((..........)))))))..(((((((....)))))))......... ( -42.20) >consensus G_C_CGUCCUCAUCACUCAGCAGAUCCGACUGCUG_UGAUGCUGCUGCUGCUGAUGGUGAUUCAAUUGCAAAUGGCGAAGUUUACGCUUGCGCGACGACGUCACCGUAGUCAGCUCAUGU ....((((.((((((.(((((((........................))))))))))))).....((((....((((.......))))...)))).)))).................... (-24.81 = -23.93 + -0.88)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:13:24 2006