
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,462,704 – 4,462,936 |
| Length | 232 |
| Max. P | 0.998549 |

| Location | 4,462,704 – 4,462,824 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.89 |
| Mean single sequence MFE | -39.62 |
| Consensus MFE | -29.04 |
| Energy contribution | -29.60 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.58 |
| Structure conservation index | 0.73 |
| SVM decision value | 2.76 |
| SVM RNA-class probability | 0.996841 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4462704 120 + 22224390 AUUUCAAAUUUUACGCUCUCGCUCUCGCGGACAGCGACGCCGCCGGCGGCAGCGCAGCGCAGCCGUUGUAUUUAUAAAUACAACUUUAUAAAUACAACAUCAGUCGAUUUUCAUCUGCCG ..............(((..((((.((((.....)))).((((....)))))))).)))((((..((((((((((((((......))))))))))))))(((....)))......)))).. ( -38.40) >DroSec_CAF1 64267 120 + 1 ACUUCAAAUUUUACGCUCUCGCUCUCGCGGGCAGCGACGCCGCCGGCGGCAGCGCAGCACAGCCGUUGUAUUUAUAAAUACAACUUUAUAAAUACAACAUCAGUCGAUUUUCAUCUGCCG .............(((((((((....))))).))))..(((...)))(((((.((......)).((((((((((((((......))))))))))))))................))))). ( -39.70) >DroEre_CAF1 67969 102 + 1 ---------------C---CGCUCUCGCGGACAGCGACGCCGACGGCGGCAGCGCAGCGCAGCCGUUGUAUUUAUAAAUACAACUUUAUAAAUACAACAUCAGUCGAUUUUCAUCUGCCG ---------------(---(((....))))...(((..((((....))))..))).(.((((..((((((((((((((......))))))))))))))(((....)))......))))). ( -36.20) >DroYak_CAF1 65106 109 + 1 GCGUCGA---CUGCGC---UGCAUGCGCGAACAGCGACGCCGACGGCGGCAGCG---CGCAGC--GUGUAUUUAUAAAUACAACUUUAUAAAUACAACAUCAGUCGAUUUUCAUCUGCCG ..(((((---((((((---(((.(((((.....))...((((....)))).)))---.)))))--)((((((((((((......))))))))))))....))))))))............ ( -44.20) >consensus ACUUCAA___UUACGC___CGCUCUCGCGGACAGCGACGCCGACGGCGGCAGCGCAGCGCAGCCGUUGUAUUUAUAAAUACAACUUUAUAAAUACAACAUCAGUCGAUUUUCAUCUGCCG ..........................(((((...((((((((....)))).((........)).((((((((((((((......))))))))))))))....)))).......))))).. (-29.04 = -29.60 + 0.56)



| Location | 4,462,704 – 4,462,824 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.89 |
| Mean single sequence MFE | -45.85 |
| Consensus MFE | -36.72 |
| Energy contribution | -38.10 |
| Covariance contribution | 1.38 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.82 |
| Structure conservation index | 0.80 |
| SVM decision value | 3.14 |
| SVM RNA-class probability | 0.998549 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4462704 120 - 22224390 CGGCAGAUGAAAAUCGACUGAUGUUGUAUUUAUAAAGUUGUAUUUAUAAAUACAACGGCUGCGCUGCGCUGCCGCCGGCGGCGUCGCUGUCCGCGAGAGCGAGAGCGUAAAAUUUGAAAU (((..(((....)))..))).(((((((((((((((......)))))))))))))))..((((((.((((..(((.((((((...)))))).)))..))))..))))))........... ( -48.10) >DroSec_CAF1 64267 120 - 1 CGGCAGAUGAAAAUCGACUGAUGUUGUAUUUAUAAAGUUGUAUUUAUAAAUACAACGGCUGUGCUGCGCUGCCGCCGGCGGCGUCGCUGCCCGCGAGAGCGAGAGCGUAAAAUUUGAAGU (((..(((....)))..)))..((((((((((((((......))))))))))))))(((...((.((((((((...)))))))).)).)))(((....)))................... ( -47.70) >DroEre_CAF1 67969 102 - 1 CGGCAGAUGAAAAUCGACUGAUGUUGUAUUUAUAAAGUUGUAUUUAUAAAUACAACGGCUGCGCUGCGCUGCCGCCGUCGGCGUCGCUGUCCGCGAGAGCG---G--------------- ((((.(((....)))(((....((((((((((((((......))))))))))))))(((.(((...))).)))(((...)))))))))).((((....)))---)--------------- ( -44.40) >DroYak_CAF1 65106 109 - 1 CGGCAGAUGAAAAUCGACUGAUGUUGUAUUUAUAAAGUUGUAUUUAUAAAUACAC--GCUGCG---CGCUGCCGCCGUCGGCGUCGCUGUUCGCGCAUGCA---GCGCAG---UCGACGC ..((.(((....)))(((((....((((((((((((......))))))))))))(--((((((---((.(((((....))))).)))(((....))).)))---))))))---))...)) ( -43.20) >consensus CGGCAGAUGAAAAUCGACUGAUGUUGUAUUUAUAAAGUUGUAUUUAUAAAUACAACGGCUGCGCUGCGCUGCCGCCGGCGGCGUCGCUGUCCGCGAGAGCG___GCGUAA___UUGAAGU (((..(((....)))..)))..((((((((((((((......))))))))))))))(((.(((..((((((((...))))))))))).)))(((....)))................... (-36.72 = -38.10 + 1.38)


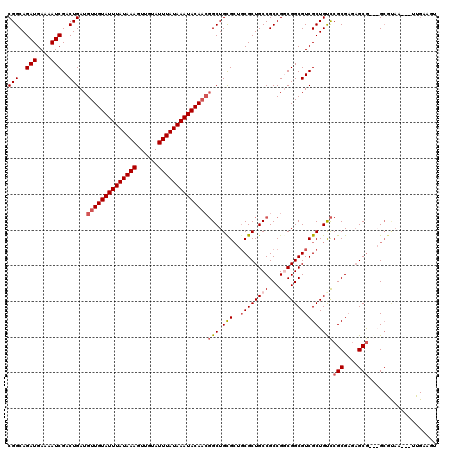
| Location | 4,462,744 – 4,462,858 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.31 |
| Mean single sequence MFE | -50.65 |
| Consensus MFE | -46.69 |
| Energy contribution | -48.25 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.00 |
| Mean z-score | -6.23 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.924449 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4462744 114 + 22224390 CGCCGGCGGCAGCGCAGCGCAGCCGUUGUAUUUAUAAAUACAACUUUAUAAAUACAACAUCAGUCGAUUUUCAUCUGCCGAAUCGGUUGCGCUGCGGCUGCGCUGAGA------AUAAGA ...(((((.(((((((((((((((((((((((((((((......)))))))))))))).((.((.(((....))).)).))...))))))))))).)))))))))...------...... ( -55.20) >DroSec_CAF1 64307 114 + 1 CGCCGGCGGCAGCGCAGCACAGCCGUUGUAUUUAUAAAUACAACUUUAUAAAUACAACAUCAGUCGAUUUUCAUCUGCCGAAUCGGUUGCGCUGCGGCUGCGCUGAGA------ACAAAA ...(((((.(((((((((.(((((((((((((((((((......)))))))))))))).((.((.(((....))).)).))...))))).))))).)))))))))...------...... ( -49.80) >DroEre_CAF1 67991 114 + 1 CGACGGCGGCAGCGCAGCGCAGCCGUUGUAUUUAUAAAUACAACUUUAUAAAUACAACAUCAGUCGAUUUUCAUCUGCCGAAUCGGUUGCGCUGCGGCUGCGCUGAGA------ACAAGA ...(((((.(((((((((((((((((((((((((((((......)))))))))))))).((.((.(((....))).)).))...))))))))))).)))))))))...------...... ( -55.40) >DroYak_CAF1 65140 113 + 1 CGACGGCGGCAGCG---CGCAGC--GUGUAUUUAUAAAUACAACUUUAUAAAUACAACAUCAGUCGAUUUUCAUCUGCCGAAU--GUUGCGUUGCGGCUGCGCUGAUAGAAAAUACAAGA ...(((((.((((.---((((((--(((((((((((((......)))))))))))((((((.((.(((....))).)).).))--))))))))))))))))))))............... ( -42.20) >consensus CGACGGCGGCAGCGCAGCGCAGCCGUUGUAUUUAUAAAUACAACUUUAUAAAUACAACAUCAGUCGAUUUUCAUCUGCCGAAUCGGUUGCGCUGCGGCUGCGCUGAGA______ACAAGA ...(((((.(((((((((((((((((((((((((((((......)))))))))))))).((.((.(((....))).)).))...))))))))))).)))))))))............... (-46.69 = -48.25 + 1.56)

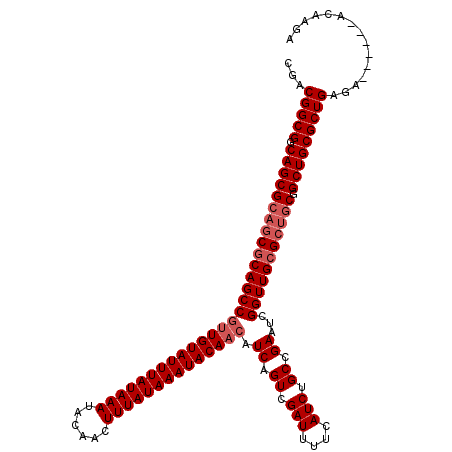
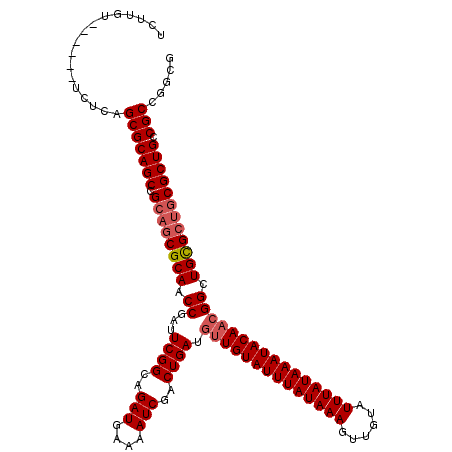
| Location | 4,462,744 – 4,462,858 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.31 |
| Mean single sequence MFE | -48.98 |
| Consensus MFE | -45.88 |
| Energy contribution | -47.00 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.03 |
| Mean z-score | -5.55 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.966565 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4462744 114 - 22224390 UCUUAU------UCUCAGCGCAGCCGCAGCGCAACCGAUUCGGCAGAUGAAAAUCGACUGAUGUUGUAUUUAUAAAGUUGUAUUUAUAAAUACAACGGCUGCGCUGCGCUGCCGCCGGCG ......------..((.(((((((.((((((((.((...((((..(((....)))..)))).((((((((((((((......)))))))))))))))).)))))))))))).))).)).. ( -53.20) >DroSec_CAF1 64307 114 - 1 UUUUGU------UCUCAGCGCAGCCGCAGCGCAACCGAUUCGGCAGAUGAAAAUCGACUGAUGUUGUAUUUAUAAAGUUGUAUUUAUAAAUACAACGGCUGUGCUGCGCUGCCGCCGGCG ......------..((.(((((((.((((((((.((...((((..(((....)))..)))).((((((((((((((......)))))))))))))))).)))))))))))).))).)).. ( -51.30) >DroEre_CAF1 67991 114 - 1 UCUUGU------UCUCAGCGCAGCCGCAGCGCAACCGAUUCGGCAGAUGAAAAUCGACUGAUGUUGUAUUUAUAAAGUUGUAUUUAUAAAUACAACGGCUGCGCUGCGCUGCCGCCGUCG ......------.....(((((((.((((((((.((...((((..(((....)))..)))).((((((((((((((......)))))))))))))))).)))))))))))).)))..... ( -53.00) >DroYak_CAF1 65140 113 - 1 UCUUGUAUUUUCUAUCAGCGCAGCCGCAACGCAAC--AUUCGGCAGAUGAAAAUCGACUGAUGUUGUAUUUAUAAAGUUGUAUUUAUAAAUACAC--GCUGCG---CGCUGCCGCCGUCG .................(((((((((((.((.(((--((.(((..(((....)))..))))))))(((((((((((......))))))))))).)--).))))---.)))).)))..... ( -38.40) >consensus UCUUGU______UCUCAGCGCAGCCGCAGCGCAACCGAUUCGGCAGAUGAAAAUCGACUGAUGUUGUAUUUAUAAAGUUGUAUUUAUAAAUACAACGGCUGCGCUGCGCUGCCGCCGGCG .................(((((((.((((((((.((...((((..(((....)))..)))).((((((((((((((......)))))))))))))))).)))))))))))).)))..... (-45.88 = -47.00 + 1.12)


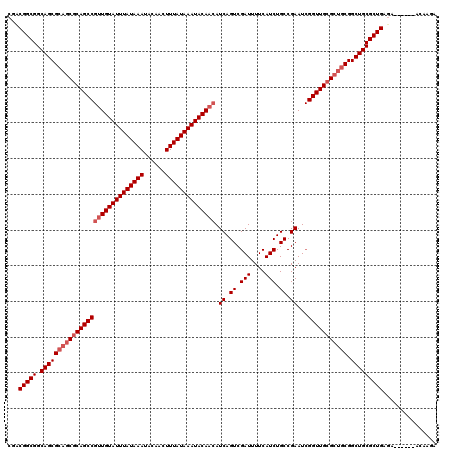
| Location | 4,462,824 – 4,462,936 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.73 |
| Mean single sequence MFE | -24.98 |
| Consensus MFE | -21.31 |
| Energy contribution | -21.88 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.781234 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4462824 112 + 22224390 AAUCGGUUGCGCUGCGGCUGCGCUGAGA------AUAAGAUACAAAACUCGUGCGCGGCGCUUCAAGAAACCAACUCAAAAGCAGACAAAUCAAAUAACUAA--CUUGUAGAAAAUAAAA ....((((...((((.(((((((((((.------.............)))).))))))).......((.......))....))))...))))..........--................ ( -25.34) >DroSec_CAF1 64387 112 + 1 AAUCGGUUGCGCUGCGGCUGCGCUGAGA------ACAAAAUACAAAACUCGUGCGCGGCGCUUCAAGAAACCAACUCAAAAGCAGACAAAUCAAAUAACUAA--CUUGUAGAAAAUAAAA ....((((...((((.(((((((((((.------.............)))).))))))).......((.......))....))))...))))..........--................ ( -25.34) >DroEre_CAF1 68071 108 + 1 AAUCGGUUGCGCUGCGGCUGCGCUGAGA------ACAAGAUACAAAACUCGUGCGCGGCGCUUCAAGUAACCAACUCAUAAGCAGACAAAUCAAAUAAUUAA--CUUGUCGAAAAA---- ..((((((((...(((.((((((((((.------.............)))).))))))))).....))))))............(((((.............--.)))))))....---- ( -30.48) >DroYak_CAF1 65215 114 + 1 AAU--GUUGCGUUGCGGCUGCGCUGAUAGAAAAUACAAGAUACAAAACUCGUGCGCGGCGCUUCAAGAAACCAACUCAAAAGCAGACAAAUCAAAUAUCUAAUUCUUGUGGAAAAU---- ..(--((((((..(((.(((((((((......................))).)))))))))..)..((.......))....))).))).........((((.......))))....---- ( -18.75) >consensus AAUCGGUUGCGCUGCGGCUGCGCUGAGA______ACAAGAUACAAAACUCGUGCGCGGCGCUUCAAGAAACCAACUCAAAAGCAGACAAAUCAAAUAACUAA__CUUGUAGAAAAU____ ....((((...((((.(((((((((((....................)))).))))))).......((.......))....))))...))))............................ (-21.31 = -21.88 + 0.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:13:19 2006