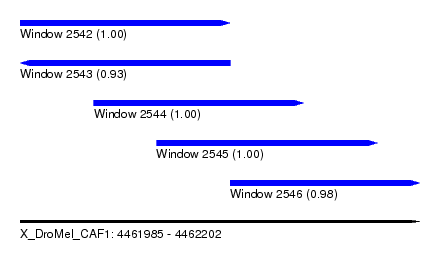
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,461,985 – 4,462,202 |
| Length | 217 |
| Max. P | 0.999359 |

| Location | 4,461,985 – 4,462,099 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 95.32 |
| Mean single sequence MFE | -41.73 |
| Consensus MFE | -41.55 |
| Energy contribution | -41.00 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.34 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.59 |
| SVM RNA-class probability | 0.995562 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
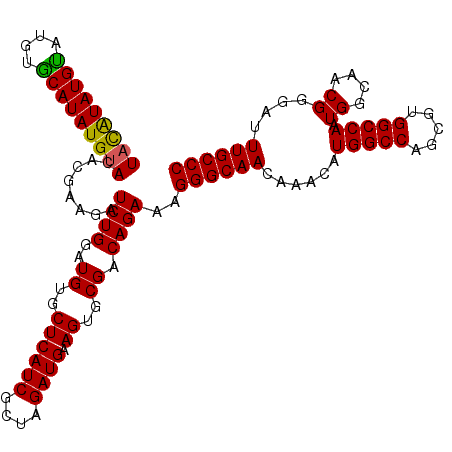
>X_DroMel_CAF1 4461985 114 + 22224390 UACAUAUGUAUGGGCAUAUGUACACGAAGAUCUGGAUGUGCUCAUCGCUAGAUGAAGUGCGACAGAAAGGGCAACAAACAUGGCCAGCGUGGCCAAUGGCAACGGGAUUUGCCC (((((((((....)))))))))........((((..((..((((((....)))).))..)).))))..((((((......(((((.....))))).((....))....)))))) ( -42.30) >DroSim_CAF1 68991 114 + 1 UACAUAUGCAUGUGCAUAUGUACACAAUGAUCUGGAUGUGCUCAUCGCUAGAUGAAGUGCGACAGAAAGGGCAACAAACAUGGCCAGCGUGGCCAAUGGCAACGGGAUUUGCCC (((((((((....)))))))))........((((..((..((((((....)))).))..)).))))..((((((......(((((.....))))).((....))....)))))) ( -44.40) >DroEre_CAF1 67206 114 + 1 UAUGUAUGUAUGUACAUAUGCACACGAAGAUCUGGAUGUGCUCAUCGCUAGAUGAAGUGCGACAGAAAGGGCAACAAACAUGGCCAGCGUGGCCAAUGGCAACGGGAUUUGCCC ..(((.((((((....)))))).)))....((((..((..((((((....)))).))..)).))))..((((((......(((((.....))))).((....))....)))))) ( -38.50) >consensus UACAUAUGUAUGUGCAUAUGUACACGAAGAUCUGGAUGUGCUCAUCGCUAGAUGAAGUGCGACAGAAAGGGCAACAAACAUGGCCAGCGUGGCCAAUGGCAACGGGAUUUGCCC (((((((((....)))))))))........((((..((..((((((....)))).))..)).))))..((((((......(((((.....))))).((....))....)))))) (-41.55 = -41.00 + -0.55)

| Location | 4,461,985 – 4,462,099 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 95.32 |
| Mean single sequence MFE | -35.23 |
| Consensus MFE | -32.91 |
| Energy contribution | -33.47 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933724 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
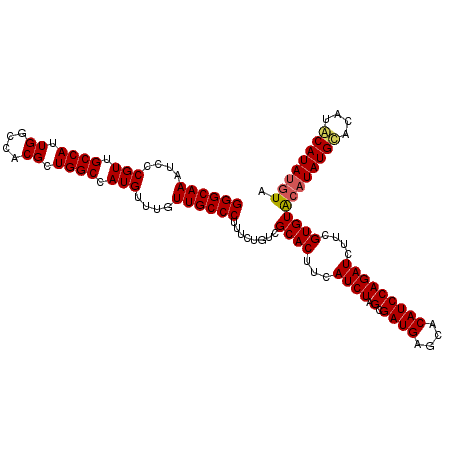
>X_DroMel_CAF1 4461985 114 - 22224390 GGGCAAAUCCCGUUGCCAUUGGCCACGCUGGCCAUGUUUGUUGCCCUUUCUGUCGCACUUCAUCUAGCGAUGAGCACAUCCAGAUCUUCGUGUACAUAUGCCCAUACAUAUGUA ((((((....(((.((((.((....)).)))).)))....))))))..((((......((((((....))))))......))))........((((((((......)))))))) ( -33.80) >DroSim_CAF1 68991 114 - 1 GGGCAAAUCCCGUUGCCAUUGGCCACGCUGGCCAUGUUUGUUGCCCUUUCUGUCGCACUUCAUCUAGCGAUGAGCACAUCCAGAUCAUUGUGUACAUAUGCACAUGCAUAUGUA ((((((....(((.((((.((....)).)))).)))....))))))..((.(((((..........)))))))(((((..........)))))((((((((....)))))))). ( -38.30) >DroEre_CAF1 67206 114 - 1 GGGCAAAUCCCGUUGCCAUUGGCCACGCUGGCCAUGUUUGUUGCCCUUUCUGUCGCACUUCAUCUAGCGAUGAGCACAUCCAGAUCUUCGUGUGCAUAUGUACAUACAUACAUA ((((((....(((.((((.((....)).)))).)))....))))))..((.(((((..........)))))))((((((..........)))))).((((((......)))))) ( -33.60) >consensus GGGCAAAUCCCGUUGCCAUUGGCCACGCUGGCCAUGUUUGUUGCCCUUUCUGUCGCACUUCAUCUAGCGAUGAGCACAUCCAGAUCUUCGUGUACAUAUGCACAUACAUAUGUA ((((((....(((.((((.((....)).)))).)))....))))))........((((...((((.(.((((....)))))))))....))))((((((((....)))))))). (-32.91 = -33.47 + 0.56)


| Location | 4,462,025 – 4,462,139 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.85 |
| Mean single sequence MFE | -34.16 |
| Consensus MFE | -31.62 |
| Energy contribution | -32.10 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.93 |
| SVM decision value | 3.54 |
| SVM RNA-class probability | 0.999359 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4462025 114 + 22224390 CUCAUCGCUAGAUGA------AGUGCGACAGAAAGGGCAACAAACAUGGCCAGCGUGGCCAAUGGCAACGGGAUUUGCCCAAAAAUCCAAAAUGCAUCCAGACAUAUGCAUAUGUGUAUG .(((((....)))))------.((((.(((....((((((......(((((.....))))).((....))....))))))...........((((((........)))))).))))))). ( -34.40) >DroSec_CAF1 63560 114 + 1 CUCAUCGCUAGAUGA------AGUGCGACAGAAAGGGCAACAAACAUGGCCAGCGUGGCCAAUGGCAACGGGAUCUGCCCAAAAAUCCAAAAUGCAUCCAGACAUAUGCGUAUGUGUAUG .(((((....)))))------.((((.(((....(((((.......(((((.....))))).((....)).....)))))...........((((((........)))))).))))))). ( -33.30) >DroSim_CAF1 69031 114 + 1 CUCAUCGCUAGAUGA------AGUGCGACAGAAAGGGCAACAAACAUGGCCAGCGUGGCCAAUGGCAACGGGAUUUGCCCAAAAAUCCAAAACGCAUCCAGACAUAUGCAUAUGUGUAUG .(((((....)))))------.((((.(((....((((((......(((((.....))))).((....))....)))))).............((((........))))...))))))). ( -32.70) >DroEre_CAF1 67246 114 + 1 CUCAUCGCUAGAUGA------AGUGCGACAGAAAGGGCAACAAACAUGGCCAGCGUGGCCAAUGGCAACGGGAUUUGCCCAAAAAUCCAAAAUGCAUCCAGACAUAUGCAUAUGUGUAUG .(((((....)))))------.((((.(((....((((((......(((((.....))))).((....))....))))))...........((((((........)))))).))))))). ( -34.40) >DroYak_CAF1 64419 118 + 1 --CAUCGCUAGAUGAUGUGCAAGUGCGACAGAAAGGGCAACAAACAUGGCCAGCGUGGCCAAUGGCAACGGGAUUUGCCCGAAAAUCCAAAAUGCAUCCAGGAAAAUGCAUAUGUGUAUG --((((....))))(((..((.(((((.......((((((......(((((.....))))).((....))....)))))).....(((............)))...))))).))..))). ( -36.00) >consensus CUCAUCGCUAGAUGA______AGUGCGACAGAAAGGGCAACAAACAUGGCCAGCGUGGCCAAUGGCAACGGGAUUUGCCCAAAAAUCCAAAAUGCAUCCAGACAUAUGCAUAUGUGUAUG .(((((....))))).......((((.(((....((((((......(((((.....))))).((....))....))))))...........((((((........)))))).))))))). (-31.62 = -32.10 + 0.48)


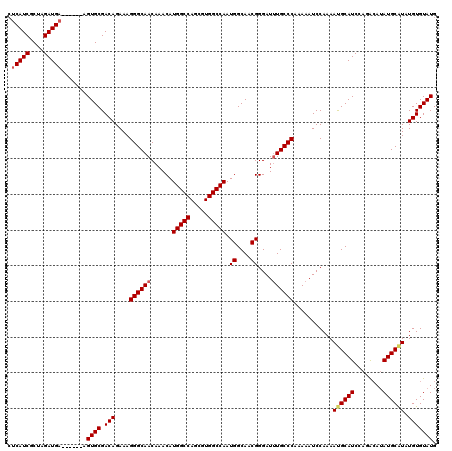
| Location | 4,462,059 – 4,462,179 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.21 |
| Mean single sequence MFE | -39.42 |
| Consensus MFE | -32.74 |
| Energy contribution | -33.18 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.89 |
| Structure conservation index | 0.83 |
| SVM decision value | 3.14 |
| SVM RNA-class probability | 0.998551 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4462059 120 + 22224390 CAAACAUGGCCAGCGUGGCCAAUGGCAACGGGAUUUGCCCAAAAAUCCAAAAUGCAUCCAGACAUAUGCAUAUGUGUAUGUGGGUGCUACAUACAGUGAUCACUCGUAUAGAAGGCAGAA ......(((((.....))))).((....))...((((((..............((((((..((((((((....))))))))))))))...(((((((....))).))))....)))))). ( -37.30) >DroSec_CAF1 63594 120 + 1 CAAACAUGGCCAGCGUGGCCAAUGGCAACGGGAUCUGCCCAAAAAUCCAAAAUGCAUCCAGACAUAUGCGUAUGUGUAUGUGGGUGCUACAUACAGUGAUCACUUGUAUGAAAGGCAUGA ......(((((.....)))))(((.(...(((.....))).............((((((..((((((((....))))))))))))))..((((((((....)).))))))...).))).. ( -37.80) >DroSim_CAF1 69065 120 + 1 CAAACAUGGCCAGCGUGGCCAAUGGCAACGGGAUUUGCCCAAAAAUCCAAAACGCAUCCAGACAUAUGCAUAUGUGUAUGUGGGUGCUACAUACAGUGAUCACUCGUAUGAAAGGCAGGA ....(((((((.....)))).)))((....((((((......)))))).....((((((..((((((((....))))))))))))))..((((((((....))).)))))....)).... ( -40.50) >DroEre_CAF1 67280 115 + 1 CAAACAUGGCCAGCGUGGCCAAUGGCAACGGGAUUUGCCCAAAAAUCCAAAAUGCAUCCAGACAUAUGCAUAUGUGUAUGUGGGUGCUAUAUACAGUGGGCACUCGUGGCGCGG-----A ....(((((((.....)))).)))((.(((((...((((((............((((((..((((((((....)))))))))))))).........))))))))))).))....-----. ( -43.40) >DroYak_CAF1 64457 115 + 1 CAAACAUGGCCAGCGUGGCCAAUGGCAACGGGAUUUGCCCGAAAAUCCAAAAUGCAUCCAGGAAAAUGCAUAUGUGUAUGUGGGUGCUACAUACAGUGUGCACUCGUAGCACAG-----A ....(((((((.....)))).)))((.(((((((((......))))))..................(((((((.(((((((((...))))))))))))))))..))).))....-----. ( -38.10) >consensus CAAACAUGGCCAGCGUGGCCAAUGGCAACGGGAUUUGCCCAAAAAUCCAAAAUGCAUCCAGACAUAUGCAUAUGUGUAUGUGGGUGCUACAUACAGUGAUCACUCGUAUCAAAGGCA__A ......(((((.....))))).((....))((((((......)))))).....(((((((..(((((((....))))))))))))))....((((((....))).)))............ (-32.74 = -33.18 + 0.44)

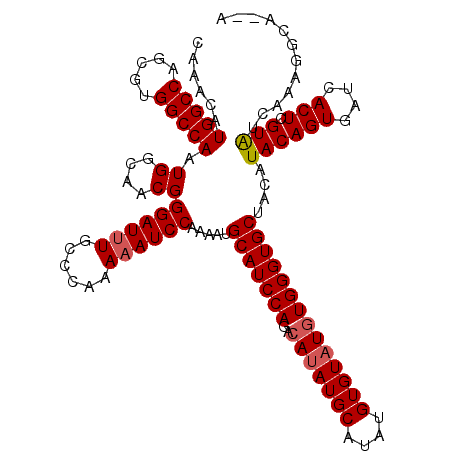
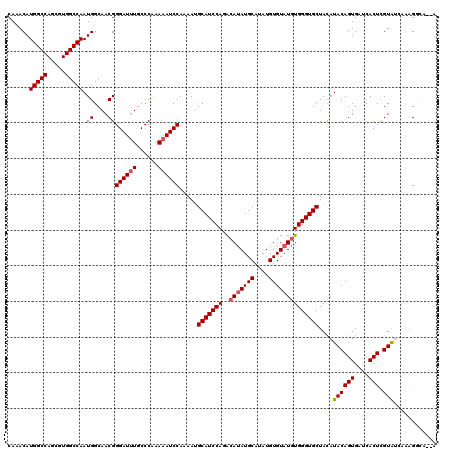
| Location | 4,462,099 – 4,462,202 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.21 |
| Mean single sequence MFE | -27.66 |
| Consensus MFE | -17.42 |
| Energy contribution | -18.66 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.978853 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4462099 103 + 22224390 AAAAAUCCAAAAUGCAUCCAGACAUAUGCAUAUGUGUAUGUGGGUGCUACAUACAGUGAUCACUCGUAUAGAAGGCAGAAAAAGU-UGACAAU------------UGGAGAUUUGG---- .....(((((...((((((..((((((((....))))))))))))))...(((((((....))).))))................-......)------------)))).......---- ( -24.70) >DroSec_CAF1 63634 103 + 1 AAAAAUCCAAAAUGCAUCCAGACAUAUGCGUAUGUGUAUGUGGGUGCUACAUACAGUGAUCACUUGUAUGAAAGGCAUGAAAAGU-UGACAAU------------UUGAGAUUUGG---- ..(((((((((..((((((..((((((((....))))))))))))))..((((((((....)).))))))...............-......)------------))).)))))..---- ( -27.10) >DroSim_CAF1 69105 103 + 1 AAAAAUCCAAAACGCAUCCAGACAUAUGCAUAUGUGUAUGUGGGUGCUACAUACAGUGAUCACUCGUAUGAAAGGCAGGAAAAGU-UGACAAU------------UUGAGAUUUGG---- ..(((((((((..((((((..((((((((....))))))))))))))..((((((((....))).)))))...............-......)------------))).)))))..---- ( -28.10) >DroEre_CAF1 67320 115 + 1 AAAAAUCCAAAAUGCAUCCAGACAUAUGCAUAUGUGUAUGUGGGUGCUAUAUACAGUGGGCACUCGUGGCGCGG-----AAAAGUGUGAUAAUGCUCCUCUUUACUUCAGUUUUGGUGCU ......(((((((((((((..((((((((....)))))))))))))).......((.(((((...((.((((..-----....)))).))..)))))))..........))))))).... ( -34.10) >DroYak_CAF1 64497 94 + 1 GAAAAUCCAAAAUGCAUCCAGGAAAAUGCAUAUGUGUAUGUGGGUGCUACAUACAGUGUGCACUCGUAGCACAG-----ACAAGU-UGGUAAUACUGCUU-------------------- ......((((.((((((........)))))).(((((.(((((((((.((((...)))))))))))))))))).-----.....)-)))...........-------------------- ( -24.30) >consensus AAAAAUCCAAAAUGCAUCCAGACAUAUGCAUAUGUGUAUGUGGGUGCUACAUACAGUGAUCACUCGUAUCAAAGGCA__AAAAGU_UGACAAU____________UUGAGAUUUGG____ ......(((((..(((((((..(((((((....))))))))))))))....((((((....))).)))...........................................))))).... (-17.42 = -18.66 + 1.24)

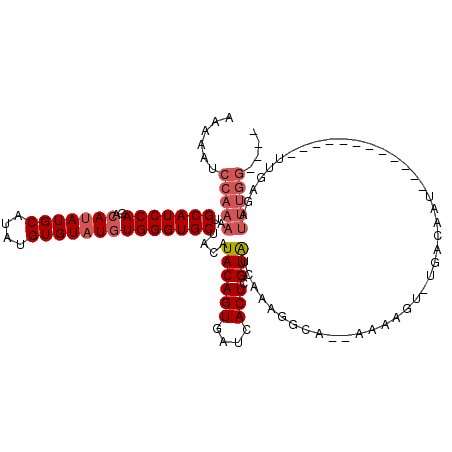

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:13:15 2006