
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,460,037 – 4,460,171 |
| Length | 134 |
| Max. P | 0.930513 |

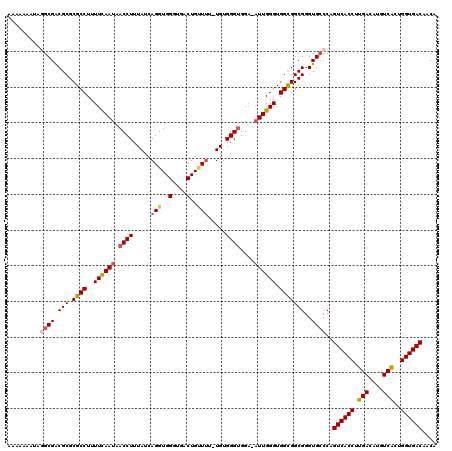
| Location | 4,460,037 – 4,460,156 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.49 |
| Mean single sequence MFE | -42.80 |
| Consensus MFE | -34.36 |
| Energy contribution | -34.88 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.909556 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4460037 119 + 22224390 AAAAAAAUAGGCGACGCGCGCCUUUUCAAUAACCUUUAUCAGGUGGGUGACUGUUUUCUGUGGGCGGA-AUUGGGUGGCGGCGGGUGCCCAGUCACCUUGACAUAUCACUGGUGACAACA .........((((.(((.((((..((((((..((((...((((.((....))....)))).))).)..-)))))).)))))))..))))..((((((.(((....)))..)))))).... ( -41.00) >DroSec_CAF1 61587 118 + 1 AAAAAAAUAGGCGACGCGCGCCUUUUCAAUAACCUUUAUCAGGUGGGUGACUGUUUU-UGUGGGUGGA-AUUGGGUGGCGGCGGGUGCCCAGUCACCUUGACAUGUCACUGGUGACAACA .........((((.(((.((((..((((((.((((.....(((..(....)..))).-...))))...-)))))).)))))))..))))..((((((.(((....)))..)))))).... ( -43.80) >DroSim_CAF1 66808 118 + 1 AAAAAAAUAGGCGACGCGCGCCUUUUCAAUAACCUUUAUCAGGUGGGUGACUGUUUU-UGUGGGUGGA-AUUGGGUGGCGGCGGGUGCCCAGUCACCUUGACAUGUCGCUGGUGACAGCA .........((((.(((.((((..((((((.((((.....(((..(....)..))).-...))))...-)))))).)))))))..))))..(((.....))).....((((....)))). ( -43.50) >DroEre_CAF1 65039 116 + 1 AAA---GAAUGCGGCGCGCGCCUUUUCAAUAACCUUUAUCAGGUGGGUGACUGGUUU-UGUGGGUGGAAAUUGGGUGGUGGCGGGUGGCCAGUCACCUUGACAUGUCACUGGUGACAACA ...---......(((((.((((..((((((.((((..(((((........)))))..-...))))....))))))....)))).)).))).((((((.(((....)))..)))))).... ( -40.00) >DroYak_CAF1 62297 112 + 1 -------AAUGCGGCGCGCGCCUUUUUAAUAACCUUUAUCAGGUGGGUGACUGCUAU-UGUGGGUGGCUUUUGGGUGGCGGCGGGUGCCCAGUCACCUUGACAUGUCACUGGUGACAACA -------..((.(((((.((((.........((((.((...((..(....)..))..-)).)))).(((.......))))))).)))))))((((((.(((....)))..)))))).... ( -45.70) >consensus AAAAAAAUAGGCGACGCGCGCCUUUUCAAUAACCUUUAUCAGGUGGGUGACUGUUUU_UGUGGGUGGA_AUUGGGUGGCGGCGGGUGCCCAGUCACCUUGACAUGUCACUGGUGACAACA .........((((.(((.((((..((((((.((((....(((........)))........))))....)))))).)))))))..))))..((((((.(((....)))..)))))).... (-34.36 = -34.88 + 0.52)


| Location | 4,460,037 – 4,460,156 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.49 |
| Mean single sequence MFE | -31.96 |
| Consensus MFE | -25.34 |
| Energy contribution | -26.38 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.660678 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4460037 119 - 22224390 UGUUGUCACCAGUGAUAUGUCAAGGUGACUGGGCACCCGCCGCCACCCAAU-UCCGCCCACAGAAAACAGUCACCCACCUGAUAAAGGUUAUUGAAAAGGCGCGCGUCGCCUAUUUUUUU ....((((((..(((....))).))))))(((((...(((((((......(-((........))).....(((...((((.....))))...)))...)))).)))..)))))....... ( -34.20) >DroSec_CAF1 61587 118 - 1 UGUUGUCACCAGUGACAUGUCAAGGUGACUGGGCACCCGCCGCCACCCAAU-UCCACCCACA-AAAACAGUCACCCACCUGAUAAAGGUUAUUGAAAAGGCGCGCGUCGCCUAUUUUUUU ...(((((....)))))..(((((((((((((((....)))(.....)...-..........-....)))))))).((((.....))))..))))..(((((.....)))))........ ( -32.80) >DroSim_CAF1 66808 118 - 1 UGCUGUCACCAGCGACAUGUCAAGGUGACUGGGCACCCGCCGCCACCCAAU-UCCACCCACA-AAAACAGUCACCCACCUGAUAAAGGUUAUUGAAAAGGCGCGCGUCGCCUAUUUUUUU .((((....))))......(((((((((((((((....)))(.....)...-..........-....)))))))).((((.....))))..))))..(((((.....)))))........ ( -32.50) >DroEre_CAF1 65039 116 - 1 UGUUGUCACCAGUGACAUGUCAAGGUGACUGGCCACCCGCCACCACCCAAUUUCCACCCACA-AAACCAGUCACCCACCUGAUAAAGGUUAUUGAAAAGGCGCGCGCCGCAUUC---UUU ...(((((....)))))..(((((((((((((..............................-...))))))))).((((.....))))..))))...(((....)))......---... ( -29.21) >DroYak_CAF1 62297 112 - 1 UGUUGUCACCAGUGACAUGUCAAGGUGACUGGGCACCCGCCGCCACCCAAAAGCCACCCACA-AUAGCAGUCACCCACCUGAUAAAGGUUAUUAAAAAGGCGCGCGCCGCAUU------- ((.(((((....)))))...)).(((((((((((....)))((.........))........-....)))))))).((((.....)))).........(((....))).....------- ( -31.10) >consensus UGUUGUCACCAGUGACAUGUCAAGGUGACUGGGCACCCGCCGCCACCCAAU_UCCACCCACA_AAAACAGUCACCCACCUGAUAAAGGUUAUUGAAAAGGCGCGCGUCGCCUAUUUUUUU ...(((((....)))))..(((((((((((((((....)))(.....)...................)))))))).((((.....))))..))))..(((((.....)))))........ (-25.34 = -26.38 + 1.04)

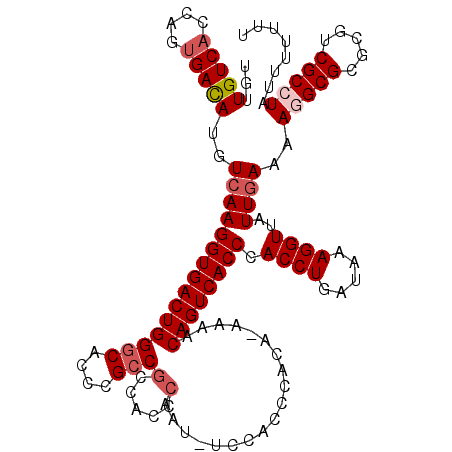
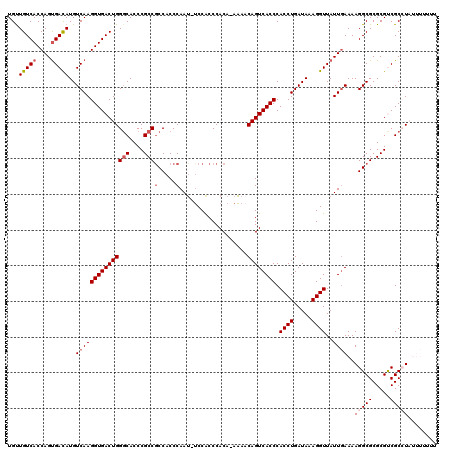
| Location | 4,460,077 – 4,460,171 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 90.10 |
| Mean single sequence MFE | -38.50 |
| Consensus MFE | -28.34 |
| Energy contribution | -29.02 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.87 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.897548 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4460077 94 + 22224390 AGGUGGGUGACUGUUUUCUGUGGGCGGA-AUUGGGUGGCGGCGGGUGCCCAGUCACCUUGACAUAUCACUGGUGACAACAGUAACAACAAUAAAA------- ..((..((.(((((((((((....))))-).((((((.(....).))))))((((((.(((....)))..)))))))))))).)).)).......------- ( -30.70) >DroSec_CAF1 61627 100 + 1 AGGUGGGUGACUGUUUU-UGUGGGUGGA-AUUGGGUGGCGGCGGGUGCCCAGUCACCUUGACAUGUCACUGGUGACAACAGCAACAACAAUAACACCCAUUA .((((((((..((((.(-(((.(.((..-..((((((.(....).))))))((((((.(((....)))..))))))..)).).)))).)))).)))))))). ( -39.20) >DroSim_CAF1 66848 100 + 1 AGGUGGGUGACUGUUUU-UGUGGGUGGA-AUUGGGUGGCGGCGGGUGCCCAGUCACCUUGACAUGUCGCUGGUGACAGCAGCAACAACAAUAACACCCAUUA (((..(....)..))).-.(((((((..-(((((((((((((....)))..))))))......(((.((((....)))).)))....))))..))))))).. ( -42.70) >DroEre_CAF1 65076 101 + 1 AGGUGGGUGACUGGUUU-UGUGGGUGGAAAUUGGGUGGUGGCGGGUGGCCAGUCACCUUGACAUGUCACUGGUGACAACAGCAACAACACCAACACCCAUUA .((((((((..((((.(-(((.(.((......((((((((((.....)))).)))))).....(((((....))))).)).).)))).)))).)))))))). ( -41.40) >consensus AGGUGGGUGACUGUUUU_UGUGGGUGGA_AUUGGGUGGCGGCGGGUGCCCAGUCACCUUGACAUGUCACUGGUGACAACAGCAACAACAAUAACACCCAUUA .((((((((..((((...(((..((......((((((.(....).))))))((((((.(((....)))..))))))....)).)))..)))).)))))))). (-28.34 = -29.02 + 0.69)

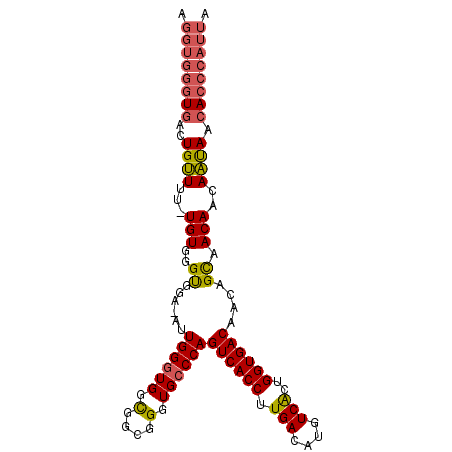

| Location | 4,460,077 – 4,460,171 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 90.10 |
| Mean single sequence MFE | -31.62 |
| Consensus MFE | -23.71 |
| Energy contribution | -24.90 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.06 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.930513 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4460077 94 - 22224390 -------UUUUAUUGUUGUUACUGUUGUCACCAGUGAUAUGUCAAGGUGACUGGGCACCCGCCGCCACCCAAU-UCCGCCCACAGAAAACAGUCACCCACCU -------.....(((.((((((((.......))))))))...)))(((((((((((....))).........(-((........)))..))))))))..... ( -26.00) >DroSec_CAF1 61627 100 - 1 UAAUGGGUGUUAUUGUUGUUGCUGUUGUCACCAGUGACAUGUCAAGGUGACUGGGCACCCGCCGCCACCCAAU-UCCACCCACA-AAAACAGUCACCCACCU ...((((((..((((..((.((....((((((..(((....))).))))))..(((....))))).)).))))-..))))))..-................. ( -32.30) >DroSim_CAF1 66848 100 - 1 UAAUGGGUGUUAUUGUUGUUGCUGCUGUCACCAGCGACAUGUCAAGGUGACUGGGCACCCGCCGCCACCCAAU-UCCACCCACA-AAAACAGUCACCCACCU ....(((((...(((.((((((((.......))))))))...)))(((((((((((....)))(.....)...-..........-....))))))))))))) ( -32.70) >DroEre_CAF1 65076 101 - 1 UAAUGGGUGUUGGUGUUGUUGCUGUUGUCACCAGUGACAUGUCAAGGUGACUGGCCACCCGCCACCACCCAAUUUCCACCCACA-AAACCAGUCACCCACCU ...(((((((((((.((((.(.((..((((((..(((....))).))))))((((.....))))............)).).)))-).))))).))))))... ( -35.50) >consensus UAAUGGGUGUUAUUGUUGUUGCUGUUGUCACCAGUGACAUGUCAAGGUGACUGGGCACCCGCCGCCACCCAAU_UCCACCCACA_AAAACAGUCACCCACCU ....(((((...(((.((((((((.......))))))))...)))(((((((((((....)))(.....)...................))))))))))))) (-23.71 = -24.90 + 1.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:13:09 2006