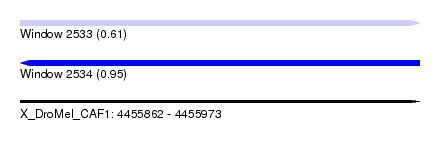
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,455,862 – 4,455,973 |
| Length | 111 |
| Max. P | 0.954997 |

| Location | 4,455,862 – 4,455,973 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.67 |
| Mean single sequence MFE | -24.92 |
| Consensus MFE | -18.34 |
| Energy contribution | -18.54 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.607444 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4455862 111 + 22224390 ACACUUGAGUAAAUAUCAACAUCAGCAACCAGCCAAGACGA---UGUCUCCAACCUCAUCGCAACGCC------UUUCUUGGCUGGUGACAAUAACCCAAAACCUAAUGAGCAAUGUAUU .....(((.......)))((((..((.((((((((((((((---((..........))))).......------..)))))))))))..........((........)).)).))))... ( -24.20) >DroSec_CAF1 57340 114 + 1 ACACUUGAGUAAAUAUCAACAUCAGCAACCAGCCAAGACGAUGAUGUCUCCAACCUCAUCGCAACGCC------UUUCUUGGCUGGUGACAAUAACCCAAAACCUAAUGAGCAACGUAUU ....((((.......)))).....((.(((((((((((((((((.((.....)).)))))).......------..)))))))))))..........((........)).))........ ( -26.70) >DroSim_CAF1 58582 114 + 1 ACACUUGAGUAAAUAUCAACAUCAGCAACCAGCCAAGACGAUGAUGUCUCCAACCUCAUCGCAACGCC------UUUCUUGGCUGGUGACAAUAACCCAAAACCUAAUGAGCAAUGUAUU .....(((.......)))((((..((.(((((((((((((((((.((.....)).)))))).......------..)))))))))))..........((........)).)).))))... ( -27.30) >DroEre_CAF1 59259 111 + 1 ACACUUGAGUAAAUAUCAACAUCAGCAACCAGCCAAGACGA---UGUCUCCAACCUCAUCGCAACGCC------UUUCUUGGCUGAUGACAAUAACCCAAAACCUAAUGAGCAAUGUAUU .....(((.......)))((((..((...((((((((((((---((..........))))).......------..)))))))))............((........)).)).))))... ( -19.60) >DroYak_CAF1 57930 117 + 1 ACACUUGAGUAAAUAUCAACAUCAGCAACCAGCCAAGACGA---UGUCUCCAACCUCAUCGCAACGCCUUUGCCUUUCUUGGCUGGUGACAAUAACCCAAAACCUAAUGAGCAAUGUAUU .....(((.......)))((((..((.(((((((((((.((---((..........))))((((.....))))...)))))))))))..........((........)).)).))))... ( -26.80) >consensus ACACUUGAGUAAAUAUCAACAUCAGCAACCAGCCAAGACGA___UGUCUCCAACCUCAUCGCAACGCC______UUUCUUGGCUGGUGACAAUAACCCAAAACCUAAUGAGCAAUGUAUU ....((((.......)))).....((.(((((((((((.((...((....))...))...((...)).........)))))))))))..........((........)).))........ (-18.34 = -18.54 + 0.20)


| Location | 4,455,862 – 4,455,973 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.67 |
| Mean single sequence MFE | -32.72 |
| Consensus MFE | -31.90 |
| Energy contribution | -31.74 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.954997 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4455862 111 - 22224390 AAUACAUUGCUCAUUAGGUUUUGGGUUAUUGUCACCAGCCAAGAAA------GGCGUUGCGAUGAGGUUGGAGACA---UCGUCUUGGCUGGUUGCUGAUGUUGAUAUUUACUCAAGUGU .((((...(((.....))).((((((...(((((((((((((((..------(((.((.....)).)))(....).---...))))))))))).((....)).))))...)))))))))) ( -30.70) >DroSec_CAF1 57340 114 - 1 AAUACGUUGCUCAUUAGGUUUUGGGUUAUUGUCACCAGCCAAGAAA------GGCGUUGCGAUGAGGUUGGAGACAUCAUCGUCUUGGCUGGUUGCUGAUGUUGAUAUUUACUCAAGUGU .((((...(((.....))).((((((...(((((((((((((((..------.((...))(((((...((....))))))).))))))))))).((....)).))))...)))))))))) ( -35.40) >DroSim_CAF1 58582 114 - 1 AAUACAUUGCUCAUUAGGUUUUGGGUUAUUGUCACCAGCCAAGAAA------GGCGUUGCGAUGAGGUUGGAGACAUCAUCGUCUUGGCUGGUUGCUGAUGUUGAUAUUUACUCAAGUGU .((((...(((.....))).((((((...(((((((((((((((..------.((...))(((((...((....))))))).))))))))))).((....)).))))...)))))))))) ( -35.40) >DroEre_CAF1 59259 111 - 1 AAUACAUUGCUCAUUAGGUUUUGGGUUAUUGUCAUCAGCCAAGAAA------GGCGUUGCGAUGAGGUUGGAGACA---UCGUCUUGGCUGGUUGCUGAUGUUGAUAUUUACUCAAGUGU .((((...(((.....))).((((((...(((((((((((((((..------(((.((.....)).)))(....).---...))))))))))).((....)).))))...)))))))))) ( -28.10) >DroYak_CAF1 57930 117 - 1 AAUACAUUGCUCAUUAGGUUUUGGGUUAUUGUCACCAGCCAAGAAAGGCAAAGGCGUUGCGAUGAGGUUGGAGACA---UCGUCUUGGCUGGUUGCUGAUGUUGAUAUUUACUCAAGUGU .((((...(((.....))).((((((...(((((((((((((((...((((.....)))).....(((.(....))---)).))))))))))).((....)).))))...)))))))))) ( -34.00) >consensus AAUACAUUGCUCAUUAGGUUUUGGGUUAUUGUCACCAGCCAAGAAA______GGCGUUGCGAUGAGGUUGGAGACA___UCGUCUUGGCUGGUUGCUGAUGUUGAUAUUUACUCAAGUGU .((((...(((.....))).((((((...(((((((((((((...........((...))(((((...((....))...)))))))))))))).((....)).))))...)))))))))) (-31.90 = -31.74 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:13:04 2006