
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,450,658 – 4,450,893 |
| Length | 235 |
| Max. P | 0.749022 |

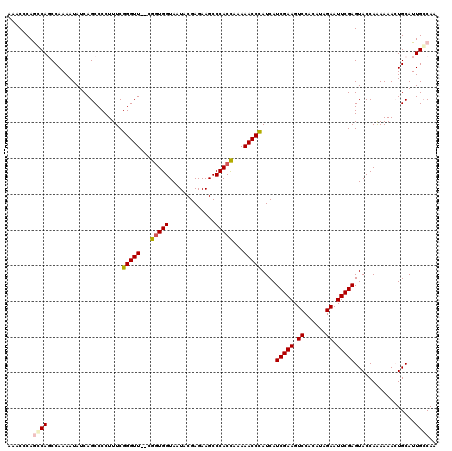
| Location | 4,450,658 – 4,450,776 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.14 |
| Mean single sequence MFE | -24.20 |
| Consensus MFE | -19.88 |
| Energy contribution | -20.40 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.617180 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
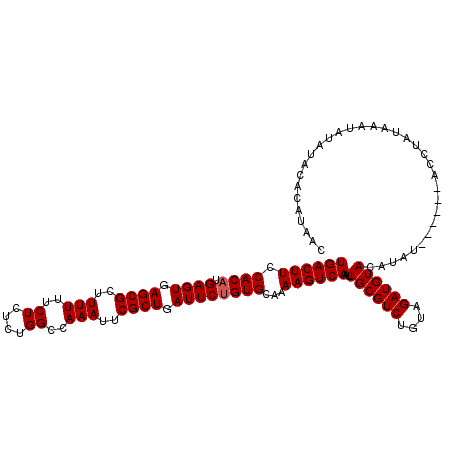
>X_DroMel_CAF1 4450658 118 - 22224390 AAACCCAGCCAGCCAAAAUAUCAGCCCCUUUCGGGUU--CGGUGGUAAUACGAGAAGCCCACCAAAAACCCAUCAUCGAAGUCCACAUAGAAUUCGAGUACCAAAAAACUGCAUUGGCAA ...........(((((.....(((........(((((--.(((((.............)))))...)))))....(((((.((......)).)))))...........)))..))))).. ( -26.12) >DroSec_CAF1 52132 120 - 1 AAACCCAGGCAGCCAAAAUAUCAGCCCCUUUCGGGUUCUCGGUGGUAAUACGAGAAGCCCACCAAAAACCCAUCAUCGAAGUCCACAUAGAAUUCGAGUACCAAAAAAGUGCAUUGCCAA .......(((((....................(((((...(((((.............)))))...)))))....(((((.((......)).)))))((((.......)))).))))).. ( -28.42) >DroSim_CAF1 53469 120 - 1 AAACCCAGGCAGCCAAAAUAUCAGCCCCUUUCGGGUUCUCGGUGGUAAUACGAGAAGCCCACCAAAAACCCAUCAUCGAAGUCCACAUAGAAUUCGAGUACCAAAAAAGUGCAUUGCCAA .......(((((....................(((((...(((((.............)))))...)))))....(((((.((......)).)))))((((.......)))).))))).. ( -28.42) >DroEre_CAF1 54114 118 - 1 AAACCCAACCAUCCAAAAUAUCAGCCCCUUUCGGGUU--CGUUGGUAAUACGAGAAGCCCACCAAAAACCCAUCAUCGAAGUCCACAUAGAAUUCGAGUACCAGAAAACUGCAUUGCCAA .......................((...(((((((((--..(((((..............))))).)))))....(((((.((......)).)))))......))))...))........ ( -19.64) >DroYak_CAF1 52802 118 - 1 AAACCCAACCAUCCAAAAUAUCAGCCCCUUUCGGGUU--CGGUGGUAAUACGAGAAGCCCACUAAAAACCUAUCAUCGAAGUCCACAUAGAAUUCGAGUACCAGAAAACUGCAUUGCCAA ...................((((((((.....)))))--.)))((((((..........................(((((.((......)).)))))....(((....))).)))))).. ( -18.40) >consensus AAACCCAGCCAGCCAAAAUAUCAGCCCCUUUCGGGUU__CGGUGGUAAUACGAGAAGCCCACCAAAAACCCAUCAUCGAAGUCCACAUAGAAUUCGAGUACCAAAAAACUGCAUUGCCAA .......((((.....................(((((...(((((.............)))))...)))))....(((((.((......)).))))).................)))).. (-19.88 = -20.40 + 0.52)


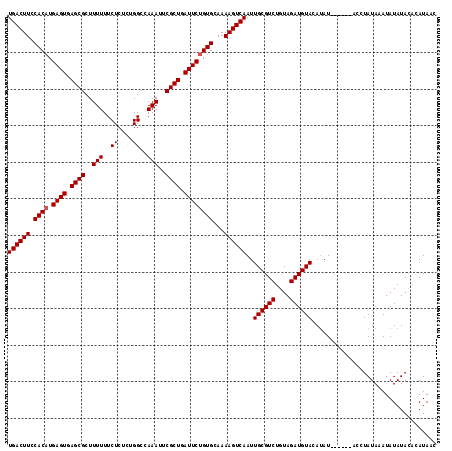
| Location | 4,450,776 – 4,450,893 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 93.04 |
| Mean single sequence MFE | -25.27 |
| Consensus MFE | -22.90 |
| Energy contribution | -23.23 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.749022 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4450776 117 + 22224390 UGACUUCCACUUGAGUGAGCGCUUUUUUCUCUCUGGCCAAAUUCGCUGAUUCUGUGCAAAAGUCAAUUGCGUCUGUAGAUGUAUAUAUAUGUACACUUAUAAAUAUAUACACAUAAC ((((((.(((..((((.((((..(((..((....))..)))..)))).)))).)))...)))))).((((....)))).(((((((((.((((....)))).)))))))))...... ( -26.60) >DroSec_CAF1 52252 111 + 1 UGACUUCCACAUGAGUGAGCGCUUUUUUCUCUCUGGCCAAAUUCGCUGAUUCUGUGCAAAAGUCAAUUGCGUCUGUAGAUGUACAUAU------ACCUAUAAAUAUAUAUAAAUAAC ((((((.((((.((((.((((..(((..((....))..)))..)))).))))))))...))))))..((((((....)))))).....------....................... ( -24.60) >DroSim_CAF1 53589 111 + 1 UGACUUCCACAUGAGUGAGCGCUUUUUUCUCUCUGGCCAAAUUCGCUGAUUCUGUGCAAAAGUCAAUUGCGUCUGUAGAUGUACACAU------ACCUAUAAAUAUAUACACAUAAC ((((((.((((.((((.((((..(((..((....))..)))..)))).))))))))...))))))..((((((....)))))).....------....................... ( -24.60) >consensus UGACUUCCACAUGAGUGAGCGCUUUUUUCUCUCUGGCCAAAUUCGCUGAUUCUGUGCAAAAGUCAAUUGCGUCUGUAGAUGUACAUAU______ACCUAUAAAUAUAUACACAUAAC ((((((.((((.((((.((((..(((..((....))..)))..)))).))))))))...))))))..((((((....)))))).................................. (-22.90 = -23.23 + 0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:12:53 2006