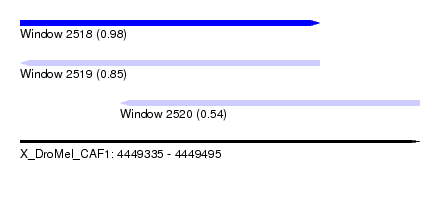
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,449,335 – 4,449,495 |
| Length | 160 |
| Max. P | 0.982806 |

| Location | 4,449,335 – 4,449,455 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.15 |
| Mean single sequence MFE | -35.08 |
| Consensus MFE | -31.67 |
| Energy contribution | -32.11 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.982806 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4449335 120 + 22224390 UCGGGCGAAGAAGUGCCCACAAAUUUGCGGUUGCUCCAUCUGGUUGCCGAGUUGAUAAAAUAAAUAUGCGGCCCGAAAAAAGGGAAGUCACAUCGGUAACAAGUAAGUGGAUGCCGCAAA ..(((((......))))).....((((((((...(((((((.((((((((..((((...............(((.......)))..))))..)))))))).))...))))).)))))))) ( -42.33) >DroSec_CAF1 50888 119 + 1 UCGGGCGAAGAAGUGCCCACAAAUUUGCGGUUGCUCCAUCUGGUCGCCGAGUUGAUAAAAUAAAUAUGCGGCCCGAAAAAAGGGAAGUCACAUCGGUAACA-GUAAGUGGAUGCCGCAAA ..(((((......))))).....((((((((...((((((((...(((((..((((...............(((.......)))..))))..)))))..))-)...))))).)))))))) ( -39.53) >DroSim_CAF1 52171 119 + 1 UCGGGCGAAGAAGUGCCCACAAAUUUGCGGUUGCUCCAUCUGGUCGCCGAGUUGAUAAAAUAAAUAUGCGGCCCGAAAAAAGGGAAGUCACAUCGGUAACA-GUAAGUGGAUGCCGCAAA ..(((((......))))).....((((((((...((((((((...(((((..((((...............(((.......)))..))))..)))))..))-)...))))).)))))))) ( -39.53) >DroEre_CAF1 52718 119 + 1 UCGGGCGAAGAAGUACCCACAAAUUUGCGGUUGCUCCAUCUGGUCACCAAGUUGAUAAAAUAAAUAUGCGGCCCGAAAAAAGGGAAGUCACAUCGGUAGCA-GUAAGUGGAUGCCACAAA ...((((.....(((.((.((....)).)).)))((((((((.(.(((..(((.....)))......((..(((.......)))..))......)))).))-)...)))))))))..... ( -25.70) >DroYak_CAF1 51459 119 + 1 UCGGGCGACGAAGUACCCACAAAUUUGCGGUUGCUCCAUCUGGUCACCGAGUUGAUAAAAUAAAUAUGCGGCCCGAAAAAUGGGAAGUUACAUCGGUAACA-GUAAGUGGAUGCCACAAA ((((((((.(.((((.((.((....)).)).)))).).))..((((......))))..............))))))..........(((((....))))).-....((((...))))... ( -28.30) >consensus UCGGGCGAAGAAGUGCCCACAAAUUUGCGGUUGCUCCAUCUGGUCGCCGAGUUGAUAAAAUAAAUAUGCGGCCCGAAAAAAGGGAAGUCACAUCGGUAACA_GUAAGUGGAUGCCGCAAA ..(((((......))))).....((((((((...((((.((.((.(((((..((((...............(((.......)))..))))..))))).)).....)))))).)))))))) (-31.67 = -32.11 + 0.44)

| Location | 4,449,335 – 4,449,455 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.15 |
| Mean single sequence MFE | -32.15 |
| Consensus MFE | -28.68 |
| Energy contribution | -28.40 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.845610 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4449335 120 - 22224390 UUUGCGGCAUCCACUUACUUGUUACCGAUGUGACUUCCCUUUUUUCGGGCCGCAUAUUUAUUUUAUCAACUCGGCAACCAGAUGGAGCAACCGCAAAUUUGUGGGCACUUCUUCGCCCGA (((((((..((((....((.(((.((((((((.(..(((.......)))..)))))..............)))).))).)).))))....)))))))....(((((........))))). ( -31.93) >DroSec_CAF1 50888 119 - 1 UUUGCGGCAUCCACUUAC-UGUUACCGAUGUGACUUCCCUUUUUUCGGGCCGCAUAUUUAUUUUAUCAACUCGGCGACCAGAUGGAGCAACCGCAAAUUUGUGGGCACUUCUUCGCCCGA (((((((..((((((...-.((..((((((((.(..(((.......)))..)))))..............))))..)).)).))))....)))))))....(((((........))))). ( -32.53) >DroSim_CAF1 52171 119 - 1 UUUGCGGCAUCCACUUAC-UGUUACCGAUGUGACUUCCCUUUUUUCGGGCCGCAUAUUUAUUUUAUCAACUCGGCGACCAGAUGGAGCAACCGCAAAUUUGUGGGCACUUCUUCGCCCGA (((((((..((((((...-.((..((((((((.(..(((.......)))..)))))..............))))..)).)).))))....)))))))....(((((........))))). ( -32.53) >DroEre_CAF1 52718 119 - 1 UUUGUGGCAUCCACUUAC-UGCUACCGAUGUGACUUCCCUUUUUUCGGGCCGCAUAUUUAUUUUAUCAACUUGGUGACCAGAUGGAGCAACCGCAAAUUUGUGGGUACUUCUUCGCCCGA ...((.(((((.......-.......))))).))..........((((((............(((((.....)))))...((.((((...(((((....)))))...)))).)))))))) ( -29.74) >DroYak_CAF1 51459 119 - 1 UUUGUGGCAUCCACUUAC-UGUUACCGAUGUAACUUCCCAUUUUUCGGGCCGCAUAUUUAUUUUAUCAACUCGGUGACCAGAUGGAGCAACCGCAAAUUUGUGGGUACUUCGUCGCCCGA .(((.(((..........-.((((((((.((.....(((.......)))..(.(((.......)))).))))))))))..(((((((...(((((....)))))...))))))))))))) ( -34.00) >consensus UUUGCGGCAUCCACUUAC_UGUUACCGAUGUGACUUCCCUUUUUUCGGGCCGCAUAUUUAUUUUAUCAACUCGGCGACCAGAUGGAGCAACCGCAAAUUUGUGGGCACUUCUUCGCCCGA (((((((..((((.............((((((....(((.......)))...))))))...........((.(....).)).))))....)))))))....(((((........))))). (-28.68 = -28.40 + -0.28)

| Location | 4,449,375 – 4,449,495 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.13 |
| Mean single sequence MFE | -31.19 |
| Consensus MFE | -23.78 |
| Energy contribution | -23.02 |
| Covariance contribution | -0.76 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.542026 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4449375 120 - 22224390 UUGGACGUAGGGUGCUCCUGAUUUGCUUAUCGCAAGUACUUUUGCGGCAUCCACUUACUUGUUACCGAUGUGACUUCCCUUUUUUCGGGCCGCAUAUUUAUUUUAUCAACUCGGCAACCA .(((..((.((((.......((((((.....)))))).....((((((............(((((....)))))...((.......))))))))..............)))).))..))) ( -27.90) >DroSec_CAF1 50928 119 - 1 UUGGGCGUAGGGUGCUGCUGAUUUGCUUAUCGCAAGUACUUUUGCGGCAUCCACUUAC-UGUUACCGAUGUGACUUCCCUUUUUUCGGGCCGCAUAUUUAUUUUAUCAACUCGGCGACCA .(((.(((.(((((((((..((((((.....))))))......)))))))))......-.......((((((.(..(((.......)))..)))))))...............))).))) ( -34.40) >DroSim_CAF1 52211 119 - 1 UUGUACGUAGGGUGCUGCUGAUUUGCUUAUCGCAAGUACUUUUGCGGCAUCCACUUAC-UGUUACCGAUGUGACUUCCCUUUUUUCGGGCCGCAUAUUUAUUUUAUCAACUCGGCGACCA ..(((.((.(((((((((..((((((.....))))))......))))))))))).)))-.((..((((((((.(..(((.......)))..)))))..............))))..)).. ( -34.43) >DroEre_CAF1 52758 119 - 1 UUGAAUGUAGGGUGCUCCCGAUUUGCUUAUCGCAAGAACAUUUGUGGCAUCCACUUAC-UGCUACCGAUGUGACUUCCCUUUUUUCGGGCCGCAUAUUUAUUUUAUCAACUUGGUGACCA .........((((((.((((((((((.....))))).(((((.((((((.........-)))))).))))).............)))))..)))).......(((((.....))))))). ( -30.60) >DroYak_CAF1 51499 119 - 1 UUGGAAGUAGGGUUCUCCCGAUUUGCUUAUCGCAAAUACUUUUGUGGCAUCCACUUAC-UGUUACCGAUGUAACUUCCCAUUUUUCGGGCCGCAUAUUUAUUUUAUCAACUCGGUGACCA ..((((((((((....)))..(((((.....))))))))))))((((...))))....-.((((((((.((.....(((.......)))..(.(((.......)))).)))))))))).. ( -28.60) >consensus UUGGACGUAGGGUGCUCCUGAUUUGCUUAUCGCAAGUACUUUUGCGGCAUCCACUUAC_UGUUACCGAUGUGACUUCCCUUUUUUCGGGCCGCAUAUUUAUUUUAUCAACUCGGCGACCA ..........(((....(((((((((.....)))))......((((((............(((((....)))))...((.......))))))))................))))..))). (-23.78 = -23.02 + -0.76)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:12:51 2006