
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,448,984 – 4,449,104 |
| Length | 120 |
| Max. P | 0.789700 |

| Location | 4,448,984 – 4,449,104 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.32 |
| Mean single sequence MFE | -33.82 |
| Consensus MFE | -34.94 |
| Energy contribution | -34.46 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.23 |
| Structure conservation index | 1.03 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.789700 |
| Prediction | RNA |
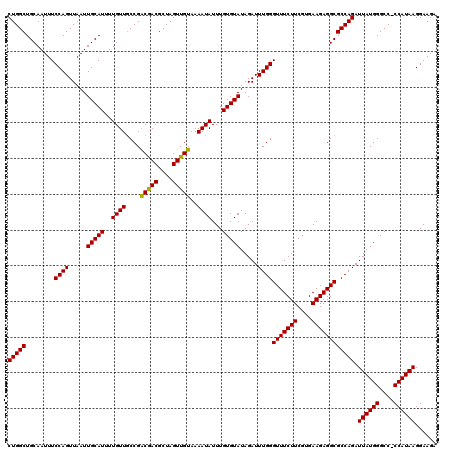
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4448984 120 + 22224390 CUGGCUGCAAUUUCCAGUUAAUUGCAUUUUGUUGCCGACAACGCUAGUUGUAAAAUAUUUGUGUAUAGAUUUGGGUUUCUUCGUGAAGAGGCGCCAGAUUAUGGGCCCCCAUAAGGAAGA (((((........((((.....(((((..((((....(((((....)))))..))))...))))).....))))(((((((....)))))))))))).((((((....))))))...... ( -36.50) >DroSec_CAF1 50529 119 + 1 CUGGCUGCAAUUUCCAGUUAAUUGCAUUUUGUUGCCGACGACGCUAGUUGUAAAAUAUUUGUGUAUAGAUUUGGGUUUCUUCGUGAAGAGGCGCCAGAUUAUGGGCC-CCAUAAGGAAGA (((((........((((.....(((((..((((....(((((....)))))..))))...))))).....))))(((((((....)))))))))))).((((((...-))))))...... ( -33.20) >DroSim_CAF1 51812 119 + 1 CUGGCUGCAAUUUCCAGUUAAUUGCAUUUUGUUGCCGACAACGCUAGUUGUAAAAUAUUUGUGUAUAGAUUUGGGUUUCUUCGUGAAGAGGCGCCAGAUUAUGGGCC-CCAUAAGGAAGA (((((........((((.....(((((..((((....(((((....)))))..))))...))))).....))))(((((((....)))))))))))).((((((...-))))))...... ( -33.50) >DroEre_CAF1 52391 119 + 1 CUGGCUGCAAUUUCCAGUUAAUUGCAUUUUGUUACCGGCGACGCUAGUUGUAAAAUAUUUGUGUAUAGAUUUGGGUUUCUUCGUGAAGAGGCGCCAGAUUAUGGGCC-CCAUAAGGAAGA (((((........((((.....(((((..((((....(((((....)))))..))))...))))).....))))(((((((....)))))))))))).((((((...-))))))...... ( -32.40) >DroYak_CAF1 51132 119 + 1 CUGGCUGCAAUUUCCAGUUAAUUGCAUUUUGUUGCCGGCGACGCUAGUUGUAAAAUAUUUGUGUAUAGAUUUGGGUUUCUUCGUGAAGAGGCGCCAGAUUAUGGGCC-CCAUAAGGAAGA (((((........((((.....(((((..((((....(((((....)))))..))))...))))).....))))(((((((....)))))))))))).((((((...-))))))...... ( -33.50) >consensus CUGGCUGCAAUUUCCAGUUAAUUGCAUUUUGUUGCCGACGACGCUAGUUGUAAAAUAUUUGUGUAUAGAUUUGGGUUUCUUCGUGAAGAGGCGCCAGAUUAUGGGCC_CCAUAAGGAAGA (((((........((((.....(((((..((((....(((((....)))))..))))...))))).....))))(((((((....)))))))))))).((((((....))))))...... (-34.94 = -34.46 + -0.48)


| Location | 4,448,984 – 4,449,104 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.32 |
| Mean single sequence MFE | -26.55 |
| Consensus MFE | -22.48 |
| Energy contribution | -22.32 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.734445 |
| Prediction | RNA |
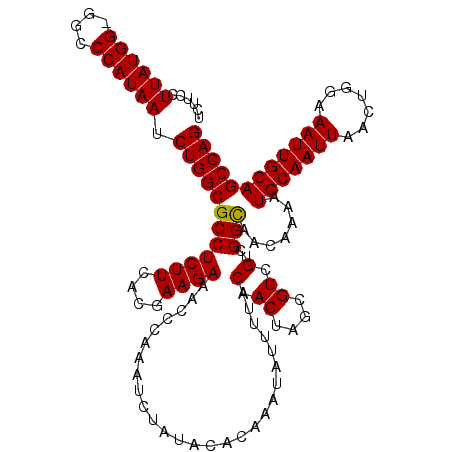
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4448984 120 - 22224390 UCUUCCUUAUGGGGGCCCAUAAUCUGGCGCCUCUUCACGAAGAAACCCAAAUCUAUACACAAAUAUUUUACAACUAGCGUUGUCGGCAACAAAAUGCAAUUAACUGGAAAUUGCAGCCAG ......((((((....)))))).((((((((((((....))))..........................(((((....))))).))).......(((((((.......)))))))))))) ( -30.20) >DroSec_CAF1 50529 119 - 1 UCUUCCUUAUGG-GGCCCAUAAUCUGGCGCCUCUUCACGAAGAAACCCAAAUCUAUACACAAAUAUUUUACAACUAGCGUCGUCGGCAACAAAAUGCAAUUAACUGGAAAUUGCAGCCAG (((((.....((-((((((.....))).))))).....)))))..............................((.((......(....)....(((((((.......))))))))).)) ( -25.40) >DroSim_CAF1 51812 119 - 1 UCUUCCUUAUGG-GGCCCAUAAUCUGGCGCCUCUUCACGAAGAAACCCAAAUCUAUACACAAAUAUUUUACAACUAGCGUUGUCGGCAACAAAAUGCAAUUAACUGGAAAUUGCAGCCAG (((((.....((-((((((.....))).))))).....)))))..........................(((((....))))).(((.......(((((((.......)))))))))).. ( -27.91) >DroEre_CAF1 52391 119 - 1 UCUUCCUUAUGG-GGCCCAUAAUCUGGCGCCUCUUCACGAAGAAACCCAAAUCUAUACACAAAUAUUUUACAACUAGCGUCGCCGGUAACAAAAUGCAAUUAACUGGAAAUUGCAGCCAG (((((.....((-((((((.....))).))))).....))))).................................(((...(((((...............)))))....)))...... ( -23.86) >DroYak_CAF1 51132 119 - 1 UCUUCCUUAUGG-GGCCCAUAAUCUGGCGCCUCUUCACGAAGAAACCCAAAUCUAUACACAAAUAUUUUACAACUAGCGUCGCCGGCAACAAAAUGCAAUUAACUGGAAAUUGCAGCCAG (((((.....((-((((((.....))).))))).....)))))..............................((.((......(....)....(((((((.......))))))))).)) ( -25.40) >consensus UCUUCCUUAUGG_GGCCCAUAAUCUGGCGCCUCUUCACGAAGAAACCCAAAUCUAUACACAAAUAUUUUACAACUAGCGUCGUCGGCAACAAAAUGCAAUUAACUGGAAAUUGCAGCCAG ......((((((....)))))).((((((((((((....))))...........................(.((....)).)..))).......(((((((.......)))))))))))) (-22.48 = -22.32 + -0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:12:49 2006