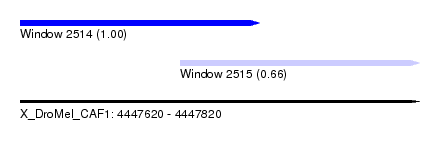
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,447,620 – 4,447,820 |
| Length | 200 |
| Max. P | 0.999695 |

| Location | 4,447,620 – 4,447,740 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.65 |
| Mean single sequence MFE | -29.48 |
| Consensus MFE | -29.02 |
| Energy contribution | -28.86 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.98 |
| SVM decision value | 3.90 |
| SVM RNA-class probability | 0.999695 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4447620 120 + 22224390 AAAUGGCCACGUCGUCGUUGCCUCAUGGCAAAAGUUCUUCUGAAUUUCUGAUGUUGCUCUGAUAAAUACUUCACUGCGCCAUUCUCGGUGGCUACCUCUCCUCCGUUCACUAUUCUUCCU .((((((.(((((....(((((....)))))((((((....))))))..))))).((..(((........)))..))))))))..(((.((........)).)))............... ( -29.80) >DroSec_CAF1 49221 119 + 1 AAAUGGCCACGUCGUCGUUGCCUCAUGGCAAAAGUUCUUCUGAAUUUCUGAUGUUGCUCUGAUAAAUACUUCACUGCGCCAUUCUCGGUGGCUCCCUCUCCUUCGCUCACUAUUCU-CCU .((((((.(((((....(((((....)))))((((((....))))))..))))).((..(((........)))..))))))))...((((((............)).)))).....-... ( -29.40) >DroSim_CAF1 50451 119 + 1 AAAUGGCCACGUCGUCGUUGCCUCAUGGCAAAAGUUCUUCUGAAUUUCUGAUGUUGCUCUGAUAAAUACUUCACUGCGCCAUUCUCGGUGGCUCCCUCUCCCUCGCUCACUAUUCU-CCU .((((((.(((((....(((((....)))))((((((....))))))..))))).((..(((........)))..))))))))...((((((............)).)))).....-... ( -29.40) >DroEre_CAF1 51075 119 + 1 AAAUGGCCACGUCGCCGUUGCCUCAUGGCAAAAGUUCUUCUGAAUUUCUGAUGUUGCUCUGAUAAGUACUUCACUGCGCCAUUCUCGGUGGCUCCCUCUCCUUCGCUCACUAUUCU-CCU .((((((.(((((....(((((....)))))((((((....))))))..))))).((..(((........)))..))))))))...((((((............)).)))).....-... ( -29.40) >DroYak_CAF1 49801 119 + 1 AAAUGGCCACGUCGUCGUUGCCUCAUGGCAAAAGUUCUUCUGAAUUUCUGAUGUUGCUCUGAUAAAUACUUCACUGCGCCAUUCUCGGUGGCUCCCUCUCCUUCGCUCACUAUUCU-CCU .((((((.(((((....(((((....)))))((((((....))))))..))))).((..(((........)))..))))))))...((((((............)).)))).....-... ( -29.40) >consensus AAAUGGCCACGUCGUCGUUGCCUCAUGGCAAAAGUUCUUCUGAAUUUCUGAUGUUGCUCUGAUAAAUACUUCACUGCGCCAUUCUCGGUGGCUCCCUCUCCUUCGCUCACUAUUCU_CCU .((((((.(((((....(((((....)))))((((((....))))))..))))).((..(((........)))..))))))))...((((((............)).))))......... (-29.02 = -28.86 + -0.16)

| Location | 4,447,700 – 4,447,820 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.48 |
| Mean single sequence MFE | -30.02 |
| Consensus MFE | -26.44 |
| Energy contribution | -26.84 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.656757 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4447700 120 + 22224390 AUUCUCGGUGGCUACCUCUCCUCCGUUCACUAUUCUUCCUCUCACUUGCUCUCUCGUUCGCAUGUUUGGGUGUAUAUCCUCAUGGAAAAUCGGAGGGGGCACAUUCGACCCACCCAUCAA ......(((((........((((((.............(((..((.(((..........))).))..)))......(((....)))....))))))((((......).)))..))))).. ( -28.90) >DroSec_CAF1 49301 119 + 1 AUUCUCGGUGGCUCCCUCUCCUUCGCUCACUAUUCU-CCUCUCACUUGCUCUCUCGUUCGCAUGUUUGGGUGUAUAUCCUCAUGGAAAAUCGGAGGGGGCACAUUCGACCCACCCAUCAA ......(((((..((((((((...((..........-..........)).......(((.((((...((((....)))).)))))))....))))))))..........)))))...... ( -28.95) >DroSim_CAF1 50531 119 + 1 AUUCUCGGUGGCUCCCUCUCCCUCGCUCACUAUUCU-CCUCUCACUUGCUCUCUCGCUCGCAUGUUUGGGUGUAUAUCCUCAUGGAAAAUCGGAGGGGGCACAUUCGACCCACCCAUCAA ....(((((((((((((((.................-.........(((.(((..((......))..))).)))..(((....))).....))))))))).)).))))............ ( -29.40) >DroEre_CAF1 51155 119 + 1 AUUCUCGGUGGCUCCCUCUCCUUCGCUCACUAUUCU-CCUCUCACUUGCUCUCUCGUUCGCAUGUUUGGGUGUAUAUCCUCAUGGAGAAUCGGUGGGGGCACAUUCGGCCCACCCAUCAA ......(((((...............((((((((((-((.......(((.(((.(((....)))...))).))).........))))))).)))))((((.......))))..))))).. ( -31.39) >DroYak_CAF1 49881 119 + 1 AUUCUCGGUGGCUCCCUCUCCUUCGCUCACUAUUCU-CCUCUCACUUGCUCUCUCGUUCGCAUGUUUGGGUGUAUAUCCACAUGGAAAAUCGGAGGGGGCACAUUCGGCCCACCCAUCAA ......(((((((((((((((...((..........-..........)).......(((.(((((..((((....))))))))))))....)))))))).......)).)))))...... ( -31.46) >consensus AUUCUCGGUGGCUCCCUCUCCUUCGCUCACUAUUCU_CCUCUCACUUGCUCUCUCGUUCGCAUGUUUGGGUGUAUAUCCUCAUGGAAAAUCGGAGGGGGCACAUUCGACCCACCCAUCAA ......(((((..((((((((.................(((..((.(((..........))).))..)))......(((....))).....))))))))..........)))))...... (-26.44 = -26.84 + 0.40)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:12:47 2006