
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,444,808 – 4,444,998 |
| Length | 190 |
| Max. P | 0.966558 |

| Location | 4,444,808 – 4,444,928 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.28 |
| Mean single sequence MFE | -38.77 |
| Consensus MFE | -30.13 |
| Energy contribution | -29.93 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.950366 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4444808 120 + 22224390 CAGAAAGUUACAGCAGCGAUAAAUUCGAGGGAAAUGUAUCUGACGAUUGUCCCUCUCACCCGGAUGGCAGAGUGGAAGGGGAACGGGAAGGGGAGGGUGCAGGUGGGGGACUAUCAGCCC ((((....((((.(..(((.....)))...)...)))))))).....((((((((((.((((.....................))))...))))))).)))(((((....)))))..... ( -35.40) >DroSec_CAF1 46453 113 + 1 CAGAAAGUUACAGCAGCGAUAAAUUCGAGGGAAAUGUAUCUGACGAUUGUCCCUCUCACCCGGAUGG-AGGGUGGAAGGG------AAAGGGGAGGGUGCAGGUGGGGGACUAUCAACCC ................(((.....))).(((...(((((((..(..((.(((((..(((((......-.)))))..))))------)))..)..)))))))(((((....)))))..))) ( -42.10) >DroSim_CAF1 47648 113 + 1 CAGAAAGUUACAGCAGCGAUAAAUUCGAGGGAAAUGUAUCUGACGAUUGUCCCUCUCACCCGGAUGG-AGGGUGGAAGGG------AAAGGGGAGGGUGCAGGUGGGGGACUAUCAACCC ................(((.....))).(((...(((((((..(..((.(((((..(((((......-.)))))..))))------)))..)..)))))))(((((....)))))..))) ( -42.10) >DroEre_CAF1 48275 109 + 1 CAGAAAGUUACAGCAGCGAUAAAUUCGAGGGAAAUGUAUCUGACGAUUGUCCCUCUCACCCGGAUGG-AUG------GUGGAGGGGGAAGGAGAGGGUGCAGGUGGGGGACUAUCA---- .............(..(((.....)))..)....(((((((........((((((((..(((.....-.))------)..))))))))......)))))))(((((....))))).---- ( -35.44) >DroYak_CAF1 47083 117 + 1 CAGAAAGUUACAGCAGCGAUAAAUUCGAGGGAAAUGUAUCUGACGAUUGUCCCUCUCACCCGGAUGG-AGAACGAAAGGGGGUGGGGAACGAGAGGGUGCAGGUGGGGGACUACCAU--C ((((....((((.(..(((.....)))...)...)))))))).....((((((((((.(((...((.-....))...)))....(....)))))))).)))(((((....)))))..--. ( -38.80) >consensus CAGAAAGUUACAGCAGCGAUAAAUUCGAGGGAAAUGUAUCUGACGAUUGUCCCUCUCACCCGGAUGG_AGGGUGGAAGGGG___GGGAAGGGGAGGGUGCAGGUGGGGGACUAUCAACCC ((((....((((.(..(((.....)))...)...)))))))).....((((((((((.(((................)))..........))))))).)))(((((....)))))..... (-30.13 = -29.93 + -0.20)


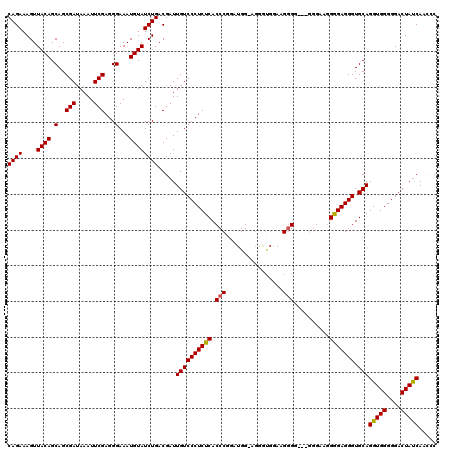
| Location | 4,444,848 – 4,444,967 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.63 |
| Mean single sequence MFE | -47.74 |
| Consensus MFE | -33.33 |
| Energy contribution | -33.13 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938312 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4444848 119 + 22224390 UGACGAUUGUCCCUCUCACCCGGAUGGCAGAGUGGAAGGGGAACGGGAAGGGGAGGGUGCAGGUGGGGGACUAUCAGCCCAA-UCCAAUCCCUCCGAGUCACCCCAUCCGACAAGGAUGA ((((.....(((((..(((.(........).)))..)))))...((((..(((..((.((.(((((....))))).))))..-)))..)))).....))))...(((((.....))))). ( -47.50) >DroSec_CAF1 46493 112 + 1 UGACGAUUGUCCCUCUCACCCGGAUGG-AGGGUGGAAGGG------AAAGGGGAGGGUGCAGGUGGGGGACUAUCAACCCAA-UCCAAUCCCUCCGAGUCACCCCAUCCGACAAGGAUGA ((((.....(((((..(((((......-.)))))..))))------)....((((((....((..(((.........)))..-.))...))))))..))))...(((((.....))))). ( -50.30) >DroSim_CAF1 47688 112 + 1 UGACGAUUGUCCCUCUCACCCGGAUGG-AGGGUGGAAGGG------AAAGGGGAGGGUGCAGGUGGGGGACUAUCAACCCAA-UCCAAUCCCACCGAGUCACCCCAUCCGACAAGGAUGA ((((.....(((((..(((((......-.)))))..))))------)..(((..((((...(((((....))))).))))..-)))...........))))...(((((.....))))). ( -49.00) >DroEre_CAF1 48315 107 + 1 UGACGAUUGUCCCUCUCACCCGGAUGG-AUG------GUGGAGGGGGAAGGAGAGGGUGCAGGUGGGGGACUAUCA------ACCCAAUCCCUCCGAGUCACCCCAUCCGACAAGGAUGA ((((......(((((.((((.......-..)------))))))))(((.(((..((((...(((((....))))).------))))..))).)))..))))...(((((.....))))). ( -48.40) >DroYak_CAF1 47123 117 + 1 UGACGAUUGUCCCUCUCACCCGGAUGG-AGAACGAAAGGGGGUGGGGAACGAGAGGGUGCAGGUGGGGGACUACCAU--CAAUCCCAAUCCCACCAAGUCACCCCAUCCGACAAGGAUGA ((((...((((((((((.(((...((.-....))...)))....(....)))))))).)))(((((((((.......--...)))....))))))..))))...(((((.....))))). ( -43.50) >consensus UGACGAUUGUCCCUCUCACCCGGAUGG_AGGGUGGAAGGGG___GGGAAGGGGAGGGUGCAGGUGGGGGACUAUCAACCCAA_UCCAAUCCCUCCGAGUCACCCCAUCCGACAAGGAUGA ((((...((((((((((.(((................)))..........))))))).)))(((((....)))))......................))))...(((((.....))))). (-33.33 = -33.13 + -0.20)

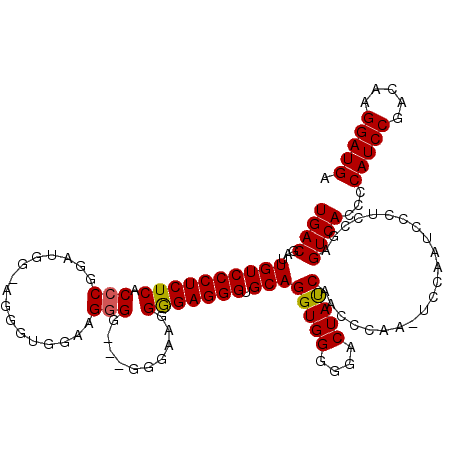
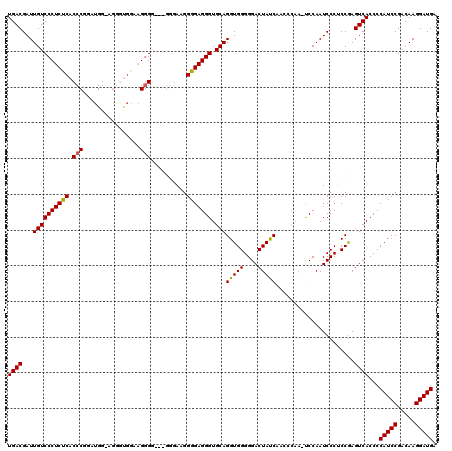
| Location | 4,444,848 – 4,444,967 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.63 |
| Mean single sequence MFE | -45.74 |
| Consensus MFE | -34.30 |
| Energy contribution | -35.22 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.966558 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4444848 119 - 22224390 UCAUCCUUGUCGGAUGGGGUGACUCGGAGGGAUUGGA-UUGGGCUGAUAGUCCCCCACCUGCACCCUCCCCUUCCCGUUCCCCUUCCACUCUGCCAUCCGGGUGAGAGGGACAAUCGUCA ......(((((((((((((......((((((...((.-..((((.....))))....))....))))))...)))))))).(((..(((((........)))))..))))))))...... ( -43.90) >DroSec_CAF1 46493 112 - 1 UCAUCCUUGUCGGAUGGGGUGACUCGGAGGGAUUGGA-UUGGGUUGAUAGUCCCCCACCUGCACCCUCCCCUUU------CCCUUCCACCCU-CCAUCCGGGUGAGAGGGACAAUCGUCA ((((((.....))))))..((((..((((((...((.-..(((........)))...))....))))))....(------((((..(((((.-......)))))..))))).....)))) ( -45.80) >DroSim_CAF1 47688 112 - 1 UCAUCCUUGUCGGAUGGGGUGACUCGGUGGGAUUGGA-UUGGGUUGAUAGUCCCCCACCUGCACCCUCCCCUUU------CCCUUCCACCCU-CCAUCCGGGUGAGAGGGACAAUCGUCA .(((((.....)))))(((((....((((((...(((-(((......))))))))))))..))))).......(------((((..(((((.-......)))))..)))))......... ( -50.20) >DroEre_CAF1 48315 107 - 1 UCAUCCUUGUCGGAUGGGGUGACUCGGAGGGAUUGGGU------UGAUAGUCCCCCACCUGCACCCUCUCCUUCCCCCUCCAC------CAU-CCAUCCGGGUGAGAGGGACAAUCGUCA ......((((((((.((((((....((.(((((((...------...))))))))).....)))))).))).....(((((((------(..-.......)))).)))))))))...... ( -43.40) >DroYak_CAF1 47123 117 - 1 UCAUCCUUGUCGGAUGGGGUGACUUGGUGGGAUUGGGAUUG--AUGGUAGUCCCCCACCUGCACCCUCUCGUUCCCCACCCCCUUUCGUUCU-CCAUCCGGGUGAGAGGGACAAUCGUCA .(((((.....)))))(((((....((((((....((((((--....))))))))))))..)))))..............((((((((..(.-......)..)))))))).......... ( -45.40) >consensus UCAUCCUUGUCGGAUGGGGUGACUCGGAGGGAUUGGA_UUGGGUUGAUAGUCCCCCACCUGCACCCUCCCCUUCCC___CCCCUUCCACCCU_CCAUCCGGGUGAGAGGGACAAUCGUCA .(((((.....)))))(((((....((.(((((((............))))))))).....)))))..............(((((.(((((........))))).))))).......... (-34.30 = -35.22 + 0.92)

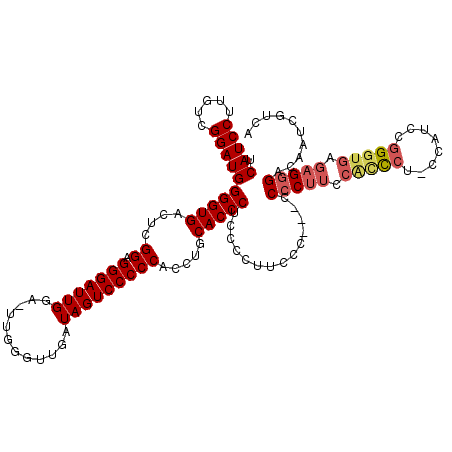

| Location | 4,444,888 – 4,444,998 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.29 |
| Mean single sequence MFE | -46.42 |
| Consensus MFE | -37.01 |
| Energy contribution | -38.25 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.80 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.507365 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4444888 110 + 22224390 GAACGGGAAGGGGAGGGUGCAGGUGGGGGACUAUCAGCCCAA-UCCAAUCCCUCCGAGUCACCCCAUCCGACAAGGAUGACCCCCCGACCCUGCGGUUGGCGUGCGGCGCG--------- ....(((..((((.((((((.((.((((((............-)))...))).))..).)))))(((((.....)))))...))))..))).((.((......)).))...--------- ( -42.90) >DroSec_CAF1 46532 113 + 1 ------AAAGGGGAGGGUGCAGGUGGGGGACUAUCAACCCAA-UCCAAUCCCUCCGAGUCACCCCAUCCGACAAGGAUGACCCCCCGACCCUGCGGUUGGCGUGCGGCGCGGCACAAAAA ------...((((.((((((.((.((((((............-)))...))).))..).)))))(((((.....))))).))))((((((....)))))).((((......))))..... ( -44.80) >DroSim_CAF1 47727 113 + 1 ------AAAGGGGAGGGUGCAGGUGGGGGACUAUCAACCCAA-UCCAAUCCCACCGAGUCACCCCAUCCGACAAGGAUGACCCCCCGACCCUGCGGUUGGCGUGCGGCGCGGCACAAAAA ------...((((.((((((.(((((((((............-)))...))))))..).)))))(((((.....))))).))))((((((....)))))).((((......))))..... ( -48.80) >DroEre_CAF1 48348 114 + 1 GAGGGGGAAGGAGAGGGUGCAGGUGGGGGACUAUCA------ACCCAAUCCCUCCGAGUCACCCCAUCCGACAAGGAUGACCCCCCGACCCUGCGGUUGGCGUGCGGCGUGGCACAAAAA ..((((((.(((..((((...(((((....))))).------))))..))).))).........(((((.....))))).))).((((((....)))))).((((......))))..... ( -48.60) >DroYak_CAF1 47162 118 + 1 GGUGGGGAACGAGAGGGUGCAGGUGGGGGACUACCAU--CAAUCCCAAUCCCACCAAGUCACCCCAUCCGACAAGGAUGACCCCCCGACCCUGCGGUUGGCGUGCGGAGCGGCACAAAAA ...((((.......((((((.(((((((((.......--...)))....))))))..).)))))(((((.....))))).))))((((((....)))))).((((......))))..... ( -47.00) >consensus G___GGGAAGGGGAGGGUGCAGGUGGGGGACUAUCAACCCAA_UCCAAUCCCUCCGAGUCACCCCAUCCGACAAGGAUGACCCCCCGACCCUGCGGUUGGCGUGCGGCGCGGCACAAAAA .........((((.(((((.(..(((((((..................)))).)))..))))))(((((.....))))).))))((((((....)))))).((((......))))..... (-37.01 = -38.25 + 1.24)

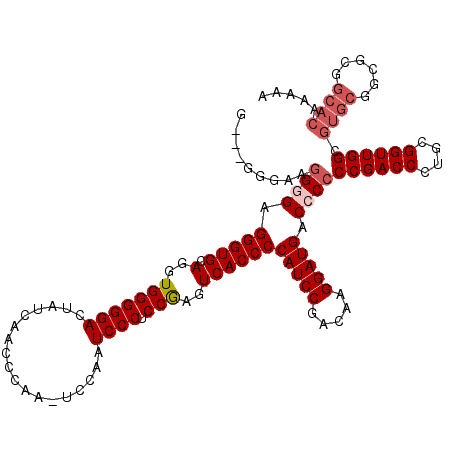
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:12:43 2006