
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,444,503 – 4,444,695 |
| Length | 192 |
| Max. P | 0.991398 |

| Location | 4,444,503 – 4,444,615 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.12 |
| Mean single sequence MFE | -41.80 |
| Consensus MFE | -28.09 |
| Energy contribution | -29.68 |
| Covariance contribution | 1.59 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.594049 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4444503 112 + 22224390 GUCUUUGGGCCCUGAGCCCUAGGCCGUGCCUUUAGCCAUCUACGGUUGCCAUCUUGACAAGACCUCGAGAG--------UCCGUAGCCCGUAGUUGGCCCAAAGCCAUUGGACACAUCAA ((((..((((.....)))).)))).(((......((((.((((((((((..((((((.......)))))).--------...)))).)))))).))))((((.....)))).)))..... ( -39.50) >DroSec_CAF1 46133 120 + 1 GUCUUUGGGCUUAGGGCCUUAGGUCUUGCCUCUAGCCAUCUACGGUUGCCAUCUUGACAAGACCUCGAGAGUCCGGGAGUCCGUAGCCCGGAGUUGGCCCAAAGCCAUUGGACACAUCAA ((((...(((((.(((((..((((((((.(..(((((......))))).......).))))))))...((.((((((.........)))))).))))))).)))))...))))....... ( -50.90) >DroSim_CAF1 47334 120 + 1 GUCUUUGGGCUUAGGGCCUUAGGCCUUGCCUCUAGCCAUCUACGGUUGCCAUCUUGACAAGACCUCGAGAGUCCGGGAGUCCGUAGCCCGGAGUUGGCCCAAAGCCAUUGGACACAUCAA ((((...(((((.(((((...(((...(((..(((....))).))).))).((((((.......)))))).((((((.........))))))...))))).)))))...))))....... ( -47.50) >DroEre_CAF1 47986 103 + 1 GUC--UGGGCCCUGAGCCUUA-------------GCCAUCUACGGUUGCCAUCUUGACAAGACCUCGAGA--CCGGGAGUCCGUAGCCUGGAGUUGGCCCAAAGCCAUUGGACACAUCAA (((--(.(((..((.(((...-------------.(((.((((((((.((.((((((.......))))))--..)).)).))))))..)))....))).))..)))...))))....... ( -37.70) >DroYak_CAF1 46775 118 + 1 GUCUUUGGGCCCUGAGCCUUAGACCUUGCCACUAGCCAUCUACGGUUGCCAUCUAGACAAGAACUCGAGA--CCAGGAGUCCGUAACCCGGAGUUGGCCCAAAGCCAUUGGACACAUCAA (.((((((((((((.(.(((((..((((...((((.((.(....).))....)))).))))..)).))).--))))((.((((.....)))).))))))))))).).............. ( -33.40) >consensus GUCUUUGGGCCCUGAGCCUUAGGCCUUGCCUCUAGCCAUCUACGGUUGCCAUCUUGACAAGACCUCGAGAG_CCGGGAGUCCGUAGCCCGGAGUUGGCCCAAAGCCAUUGGACACAUCAA (.(((((((((..(.((...((((...))))...)))..((((((...((.((((((.......)))))).....))...)))))).........))))))))).).............. (-28.09 = -29.68 + 1.59)



| Location | 4,444,503 – 4,444,615 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.12 |
| Mean single sequence MFE | -44.84 |
| Consensus MFE | -35.14 |
| Energy contribution | -37.98 |
| Covariance contribution | 2.84 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.900430 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4444503 112 - 22224390 UUGAUGUGUCCAAUGGCUUUGGGCCAACUACGGGCUACGGA--------CUCUCGAGGUCUUGUCAAGAUGGCAACCGUAGAUGGCUAAAGGCACGGCCUAGGGCUCAGGGCCCAAAGAC ....((.((((...(((((((((((..((((((.....(((--------((.....)))))((((.....)))).))))))...((.....))..)))))))))))..)))).))..... ( -43.20) >DroSec_CAF1 46133 120 - 1 UUGAUGUGUCCAAUGGCUUUGGGCCAACUCCGGGCUACGGACUCCCGGACUCUCGAGGUCUUGUCAAGAUGGCAACCGUAGAUGGCUAGAGGCAAGACCUAAGGCCCUAAGCCCAAAGAC .......(((....(((((.(((((...(((((((....)...))))))......((((((((((....((((...........))))..))))))))))..))))).)))))....))) ( -50.20) >DroSim_CAF1 47334 120 - 1 UUGAUGUGUCCAAUGGCUUUGGGCCAACUCCGGGCUACGGACUCCCGGACUCUCGAGGUCUUGUCAAGAUGGCAACCGUAGAUGGCUAGAGGCAAGGCCUAAGGCCCUAAGCCCAAAGAC .......(((....(((((.(((((...(((((((....)...))))))......((((((((((....((((...........))))..))))))))))..))))).)))))....))) ( -49.60) >DroEre_CAF1 47986 103 - 1 UUGAUGUGUCCAAUGGCUUUGGGCCAACUCCAGGCUACGGACUCCCGG--UCUCGAGGUCUUGUCAAGAUGGCAACCGUAGAUGGC-------------UAAGGCUCAGGGCCCA--GAC .......(((....(((((((((((....(((..((((((......(.--.(....)..)(((((.....))))))))))).))).-------------...)))))))))))..--))) ( -39.10) >DroYak_CAF1 46775 118 - 1 UUGAUGUGUCCAAUGGCUUUGGGCCAACUCCGGGUUACGGACUCCUGG--UCUCGAGUUCUUGUCUAGAUGGCAACCGUAGAUGGCUAGUGGCAAGGUCUAAGGCUCAGGGCCCAAAGAC ................(((((((((...((((.....))))...((((--(((..((..((((((....((((...........))))..))))))..)).)))).)))))))))))).. ( -42.10) >consensus UUGAUGUGUCCAAUGGCUUUGGGCCAACUCCGGGCUACGGACUCCCGG_CUCUCGAGGUCUUGUCAAGAUGGCAACCGUAGAUGGCUAGAGGCAAGGCCUAAGGCUCAGGGCCCAAAGAC .......(((....(((((((((((.....((((...(((....)))....))))((((((((((....((((...........))))..))))))))))..)))))))))))....))) (-35.14 = -37.98 + 2.84)

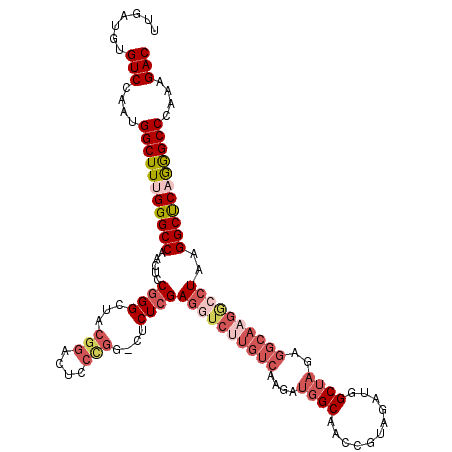
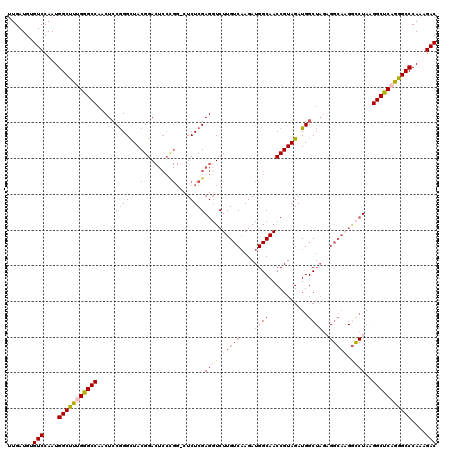
| Location | 4,444,543 – 4,444,655 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.31 |
| Mean single sequence MFE | -45.70 |
| Consensus MFE | -38.16 |
| Energy contribution | -40.24 |
| Covariance contribution | 2.08 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.27 |
| SVM RNA-class probability | 0.991398 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4444543 112 + 22224390 UACGGUUGCCAUCUUGACAAGACCUCGAGAG--------UCCGUAGCCCGUAGUUGGCCCAAAGCCAUUGGACACAUCAAUGUGGCAAUGGCAUGGCCUGCAGGAAAAUCCGGGGGGUAG (((((......((((((.......)))))).--------.)))))((((...((.((((....(((((((..((((....)))).)))))))..)))).)).((.....))...)))).. ( -42.10) >DroSec_CAF1 46173 120 + 1 UACGGUUGCCAUCUUGACAAGACCUCGAGAGUCCGGGAGUCCGUAGCCCGGAGUUGGCCCAAAGCCAUUGGACACAUCAAUGUGGCAAUGGCAUGGGCUGCAGGAAAAUCCGGGUGGUAG ......(((((((((((.......)))))).((((((..((((((((((((.(....)))...(((((((..((((....)))).)))))))..))))))).)))...))))))))))). ( -52.80) >DroSim_CAF1 47374 120 + 1 UACGGUUGCCAUCUUGACAAGACCUCGAGAGUCCGGGAGUCCGUAGCCCGGAGUUGGCCCAAAGCCAUUGGACACAUCAAUGUGGCAAUGGCAUGGGCUGCAGGAAAAUCCGGGUGGUAG ......(((((((((((.......)))))).((((((..((((((((((((.(....)))...(((((((..((((....)))).)))))))..))))))).)))...))))))))))). ( -52.80) >DroEre_CAF1 48011 118 + 1 UACGGUUGCCAUCUUGACAAGACCUCGAGA--CCGGGAGUCCGUAGCCUGGAGUUGGCCCAAAGCCAUUGGACACAUCAAUGUGGCAAAGGCAUGGUCUACAGGAAAAUGCGGGGGGUAG ......((((.(((((......(((..(((--((((((.((((.....)))).)...)))...(((.(((..((((....)))).))).)))..)))))..)))......))))))))). ( -37.20) >DroYak_CAF1 46815 118 + 1 UACGGUUGCCAUCUAGACAAGAACUCGAGA--CCAGGAGUCCGUAACCCGGAGUUGGCCCAAAGCCAUUGGACACAUCAAUGUGGCAAUGGCACGGGCUGCAGGAAAAUCCGGGGGGUAG (((((...((.(((.((.......)).)))--...))...))))).((((((.(((((((...(((((((..((((....)))).)))))))..)))))....))...))))))...... ( -43.60) >consensus UACGGUUGCCAUCUUGACAAGACCUCGAGAG_CCGGGAGUCCGUAGCCCGGAGUUGGCCCAAAGCCAUUGGACACAUCAAUGUGGCAAUGGCAUGGGCUGCAGGAAAAUCCGGGGGGUAG ......((((.((((((.......))))))..((.(((.((((((((((((.(....)))...(((((((..((((....)))).)))))))..))))))).)))...))).)).)))). (-38.16 = -40.24 + 2.08)

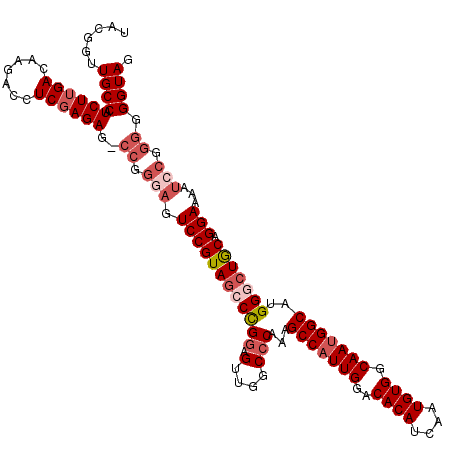

| Location | 4,444,543 – 4,444,655 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.31 |
| Mean single sequence MFE | -39.90 |
| Consensus MFE | -30.98 |
| Energy contribution | -32.62 |
| Covariance contribution | 1.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.758919 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4444543 112 - 22224390 CUACCCCCCGGAUUUUCCUGCAGGCCAUGCCAUUGCCACAUUGAUGUGUCCAAUGGCUUUGGGCCAACUACGGGCUACGGA--------CUCUCGAGGUCUUGUCAAGAUGGCAACCGUA ......(((((.....))....((((..(((((((.((((....))))..)))))))....))))......))).(((((.--------((....))...(((((.....)))))))))) ( -37.40) >DroSec_CAF1 46173 120 - 1 CUACCACCCGGAUUUUCCUGCAGCCCAUGCCAUUGCCACAUUGAUGUGUCCAAUGGCUUUGGGCCAACUCCGGGCUACGGACUCCCGGACUCUCGAGGUCUUGUCAAGAUGGCAACCGUA ......((((((......((..(((((.(((((((.((((....))))..)))))))..)))))))..)))))).(((((......(((((.....)))))((((.....)))).))))) ( -43.30) >DroSim_CAF1 47374 120 - 1 CUACCACCCGGAUUUUCCUGCAGCCCAUGCCAUUGCCACAUUGAUGUGUCCAAUGGCUUUGGGCCAACUCCGGGCUACGGACUCCCGGACUCUCGAGGUCUUGUCAAGAUGGCAACCGUA ......((((((......((..(((((.(((((((.((((....))))..)))))))..)))))))..)))))).(((((......(((((.....)))))((((.....)))).))))) ( -43.30) >DroEre_CAF1 48011 118 - 1 CUACCCCCCGCAUUUUCCUGUAGACCAUGCCUUUGCCACAUUGAUGUGUCCAAUGGCUUUGGGCCAACUCCAGGCUACGGACUCCCGG--UCUCGAGGUCUUGUCAAGAUGGCAACCGUA .........((((..((.....))..))))..((((((..((((((.((((((.....)))))))..(((.((((..(((....))))--))).))).....)))))..))))))..... ( -33.40) >DroYak_CAF1 46815 118 - 1 CUACCCCCCGGAUUUUCCUGCAGCCCGUGCCAUUGCCACAUUGAUGUGUCCAAUGGCUUUGGGCCAACUCCGGGUUACGGACUCCUGG--UCUCGAGUUCUUGUCUAGAUGGCAACCGUA ......((((((......((..(((((.(((((((.((((....))))..)))))))..)))))))..)))))).(((((((((....--....))))..(((((.....)))))))))) ( -42.10) >consensus CUACCCCCCGGAUUUUCCUGCAGCCCAUGCCAUUGCCACAUUGAUGUGUCCAAUGGCUUUGGGCCAACUCCGGGCUACGGACUCCCGG_CUCUCGAGGUCUUGUCAAGAUGGCAACCGUA ......((((((......((..(((((.(((((((.((((....))))..)))))))..)))))))..)))))).(((((.(((..........)))...(((((.....)))))))))) (-30.98 = -32.62 + 1.64)



| Location | 4,444,575 – 4,444,695 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.58 |
| Mean single sequence MFE | -42.62 |
| Consensus MFE | -38.12 |
| Energy contribution | -38.80 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.749943 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4444575 120 + 22224390 CCGUAGCCCGUAGUUGGCCCAAAGCCAUUGGACACAUCAAUGUGGCAAUGGCAUGGCCUGCAGGAAAAUCCGGGGGGUAGCAACGUAUGUGGGGUGUCUCUAUGCGACAAUUUCCUGUUU ..((.((((...((.((((....(((((((..((((....)))).)))))))..)))).)).((.....))...)))).))..((((((.(((....))))))))).............. ( -41.30) >DroSec_CAF1 46213 120 + 1 CCGUAGCCCGGAGUUGGCCCAAAGCCAUUGGACACAUCAAUGUGGCAAUGGCAUGGGCUGCAGGAAAAUCCGGGUGGUAGCAACGUAUGUGGGGUGUCUCUAUGCGACAAUUUCCUGUUU ..(((((((((.(....)))...(((((((..((((....)))).)))))))..)))))))((((((.((((.(((((....)).))).)))).((((.......)))).)))))).... ( -45.00) >DroSim_CAF1 47414 120 + 1 CCGUAGCCCGGAGUUGGCCCAAAGCCAUUGGACACAUCAAUGUGGCAAUGGCAUGGGCUGCAGGAAAAUCCGGGUGGUAGCAACGUAUGUGGGGUGUCUCUAUGCGACAAUUUCCUGUUU ..(((((((((.(....)))...(((((((..((((....)))).)))))))..)))))))((((((.((((.(((((....)).))).)))).((((.......)))).)))))).... ( -45.00) >DroEre_CAF1 48049 120 + 1 CCGUAGCCUGGAGUUGGCCCAAAGCCAUUGGACACAUCAAUGUGGCAAAGGCAUGGUCUACAGGAAAAUGCGGGGGGUAGCAACGUAUGUGGGGUGUCUCUAUGCGACAAUUUCCUGUUU ((((.((((((.(....)))...(((((..((....))...)))))..))))))))...((((((((.(((........))).((((((.(((....)))))))))....)))))))).. ( -38.10) >DroYak_CAF1 46853 120 + 1 CCGUAACCCGGAGUUGGCCCAAAGCCAUUGGACACAUCAAUGUGGCAAUGGCACGGGCUGCAGGAAAAUCCGGGGGGUAGCAACGUAUGUGGGGUGUCUCUAUGCGACAAUUUCCUGUUU (((.....)))....(((((...(((((((..((((....)))).)))))))..)))))((((((((((((...)))).....((((((.(((....)))))))))....)))))))).. ( -43.70) >consensus CCGUAGCCCGGAGUUGGCCCAAAGCCAUUGGACACAUCAAUGUGGCAAUGGCAUGGGCUGCAGGAAAAUCCGGGGGGUAGCAACGUAUGUGGGGUGUCUCUAUGCGACAAUUUCCUGUUU (((((((((((.(....)))...(((((((..((((....)))).)))))))..))))))).))......((((..((.(...((((((.(((....))))))))).).))..))))... (-38.12 = -38.80 + 0.68)



| Location | 4,444,575 – 4,444,695 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.58 |
| Mean single sequence MFE | -35.98 |
| Consensus MFE | -30.46 |
| Energy contribution | -30.94 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.85 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4444575 120 - 22224390 AAACAGGAAAUUGUCGCAUAGAGACACCCCACAUACGUUGCUACCCCCCGGAUUUUCCUGCAGGCCAUGCCAUUGCCACAUUGAUGUGUCCAAUGGCUUUGGGCCAACUACGGGCUACGG ...(((((((.((((.......)))).................((....))..)))))))..((((..(((((((.((((....))))..)))))))....)))).....((.....)). ( -35.50) >DroSec_CAF1 46213 120 - 1 AAACAGGAAAUUGUCGCAUAGAGACACCCCACAUACGUUGCUACCACCCGGAUUUUCCUGCAGCCCAUGCCAUUGCCACAUUGAUGUGUCCAAUGGCUUUGGGCCAACUCCGGGCUACGG ...(((((((.((((.......)))).................((....))..)))))))..(((((.(((((((.((((....))))..)))))))..))))).....(((.....))) ( -38.10) >DroSim_CAF1 47414 120 - 1 AAACAGGAAAUUGUCGCAUAGAGACACCCCACAUACGUUGCUACCACCCGGAUUUUCCUGCAGCCCAUGCCAUUGCCACAUUGAUGUGUCCAAUGGCUUUGGGCCAACUCCGGGCUACGG ...(((((((.((((.......)))).................((....))..)))))))..(((((.(((((((.((((....))))..)))))))..))))).....(((.....))) ( -38.10) >DroEre_CAF1 48049 120 - 1 AAACAGGAAAUUGUCGCAUAGAGACACCCCACAUACGUUGCUACCCCCCGCAUUUUCCUGUAGACCAUGCCUUUGCCACAUUGAUGUGUCCAAUGGCUUUGGGCCAACUCCAGGCUACGG ..((((((((.((((.......))))............(((........))).))))))))...(((.(((.(((.((((....))))..))).)))..)))(((.......)))..... ( -31.40) >DroYak_CAF1 46853 120 - 1 AAACAGGAAAUUGUCGCAUAGAGACACCCCACAUACGUUGCUACCCCCCGGAUUUUCCUGCAGCCCGUGCCAUUGCCACAUUGAUGUGUCCAAUGGCUUUGGGCCAACUCCGGGUUACGG ...(((((((.((((.......)))).................((....))..)))))))..(((((.(((((((.((((....))))..)))))))..))))).....(((.....))) ( -36.80) >consensus AAACAGGAAAUUGUCGCAUAGAGACACCCCACAUACGUUGCUACCCCCCGGAUUUUCCUGCAGCCCAUGCCAUUGCCACAUUGAUGUGUCCAAUGGCUUUGGGCCAACUCCGGGCUACGG .....((....((((.......))))..)).....(((.(((.......((.....))....(((((.(((((((.((((....))))..)))))))..)))))........))).))). (-30.46 = -30.94 + 0.48)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:12:40 2006