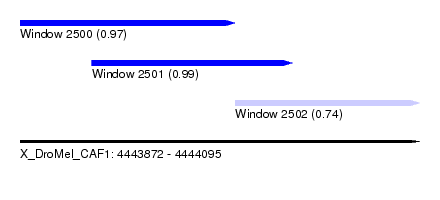
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,443,872 – 4,444,095 |
| Length | 223 |
| Max. P | 0.992909 |

| Location | 4,443,872 – 4,443,992 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.17 |
| Mean single sequence MFE | -36.60 |
| Consensus MFE | -34.62 |
| Energy contribution | -34.34 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.973980 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4443872 120 + 22224390 GAUGAAAAUGGCCCAGAGUGGGUGGAAAGUCAUAUAACAAGGACAGCGAACAGGUAGGAACUGCCUGGACAUGGCAUGCUCAUUAGGCAAAUCCUUGGAGACCCUUAUCGAGUGUGCGGA .((((...(.((((.....)))).)....))))..........((..(..(((((((...)))))))..).))((((((((..((((....((....))....))))..))))))))... ( -35.10) >DroSec_CAF1 45497 120 + 1 GAUGAAAAUGGCCCAGAGUGGGUGGAAAGUCAUAUAACAAGGACAGCGAACAGGUAGGAACUGCCUGGACAUGGCAUACUCAUUAGGCAAAUCCUUGGAGACCCUUAUCGAGUGUGCGGG .((((...(.((((.....)))).)....))))..........((..(..(((((((...)))))))..).))((((((((..((((....((....))....))))..))))))))... ( -36.10) >DroSim_CAF1 46688 120 + 1 GAUGAAAAUGGCCCAGAGUGGGUGGAAAGUCAUAUAACAAGGACAGCGAACAGGUAGGAACUGCCUGGACAUGGCAUGCUCAUUAGGCAAAUCCUUGGAGACCCUUAUCGAGUGUGCGGG .((((...(.((((.....)))).)....))))..........((..(..(((((((...)))))))..).))((((((((..((((....((....))....))))..))))))))... ( -35.10) >DroEre_CAF1 47350 120 + 1 GAUGAAAAUGGCCCAGAGUGGGUGGAAAGUCAUAUAACAAGGACAGCGAACAAGUAGGAACUGCCUGGACAUGGCAUGCUCAUUAGGCAAAUCCUUGGAGAGCCUUAUCGAGUGUGCGGG .((((...(.((((.....)))).)....))))....((.(..(((.(....(((....))).))))..).))((((((((((.((((...((....))..)))).)).))))))))... ( -34.90) >DroYak_CAF1 46161 120 + 1 GAUGAAAAUGGCCCAGAGUGGGUGGAAAGUCAUAUAAAAAGGACAACGAACAGGCAGGAACUGCCUGGACACGGCAUGCUCAUUAGGCAAAUCCUUGGAGAGCCUUAUCGAGUGUGCGGG .((((...(.((((.....)))).)....))))..............(..(((((((...)))))))..).(.((((((((((.((((...((....))..)))).)).)))))))).). ( -41.80) >consensus GAUGAAAAUGGCCCAGAGUGGGUGGAAAGUCAUAUAACAAGGACAGCGAACAGGUAGGAACUGCCUGGACAUGGCAUGCUCAUUAGGCAAAUCCUUGGAGACCCUUAUCGAGUGUGCGGG .((((...(.((((.....)))).)....))))..............(..(((((((...)))))))..).(.((((((((..((((....((....))....))))..)))))))).). (-34.62 = -34.34 + -0.28)


| Location | 4,443,912 – 4,444,024 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.25 |
| Mean single sequence MFE | -29.52 |
| Consensus MFE | -26.12 |
| Energy contribution | -26.24 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.36 |
| SVM RNA-class probability | 0.992909 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4443912 112 + 22224390 GGACAGCGAACAGGUAGGAACUGCCUGGACAUGGCAUGCUCAUUAGGCAAAUCCUUGGAGACCCUUAUCGAGUGUGCGGAGUAUAUAAAAAAAAAAAAC--------AGAAGGCGAAUAA .....((...(((((((...))))))).((.(.((((((((..((((....((....))....))))..)))))))).).)).................--------.....))...... ( -28.40) >DroSec_CAF1 45537 112 + 1 GGACAGCGAACAGGUAGGAACUGCCUGGACAUGGCAUACUCAUUAGGCAAAUCCUUGGAGACCCUUAUCGAGUGUGCGGGGGAUAUAAAAAAAAAAAAC--------AGAAGGCGAAUAA .....(((..(((((((...)))))))..)...((((((((..((((....((....))....))))..))))))))......................--------.....))...... ( -29.10) >DroSim_CAF1 46728 109 + 1 GGACAGCGAACAGGUAGGAACUGCCUGGACAUGGCAUGCUCAUUAGGCAAAUCCUUGGAGACCCUUAUCGAGUGUGCGGGGGAUAUAAAAA---AAAAC--------AGAAGGCGAAUAA .....(((..(((((((...)))))))..)...((((((((..((((....((....))....))))..))))))))..............---.....--------.....))...... ( -28.10) >DroEre_CAF1 47390 117 + 1 GGACAGCGAACAAGUAGGAACUGCCUGGACAUGGCAUGCUCAUUAGGCAAAUCCUUGGAGAGCCUUAUCGAGUGUGCGGGGGAUAUAAAAA---AAAAACAAAAAGAAAAAGGCGAAUAA .....(((..((.((((...)))).))..)...((((((((((.((((...((....))..)))).)).))))))))..............---..................))...... ( -25.30) >DroYak_CAF1 46201 109 + 1 GGACAACGAACAGGCAGGAACUGCCUGGACACGGCAUGCUCAUUAGGCAAAUCCUUGGAGAGCCUUAUCGAGUGUGCGGGGGAUAUAAAAA---AAAAC--------GGAAGUCGAAUAA .(((..((..(((((((...)))))))....(.((((((((((.((((...((....))..)))).)).)))))))).)............---....)--------)...)))...... ( -36.70) >consensus GGACAGCGAACAGGUAGGAACUGCCUGGACAUGGCAUGCUCAUUAGGCAAAUCCUUGGAGACCCUUAUCGAGUGUGCGGGGGAUAUAAAAA___AAAAC________AGAAGGCGAAUAA .....(((..(((((((...)))))))..).(.((((((((..((((....((....))....))))..)))))))).).................................))...... (-26.12 = -26.24 + 0.12)



| Location | 4,443,992 – 4,444,095 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.93 |
| Mean single sequence MFE | -23.96 |
| Consensus MFE | -19.35 |
| Energy contribution | -19.63 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.739933 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4443992 103 + 22224390 GUAUAUAAAAAAAAAAAAC--------AGAAGGCGAAUAAAACACCGAUGGGGUUGUAUUGUCACUGUCAUUGCCAGCGGAUGAGUAAUGUAGCCCCUCAUU---------CACCUUAUA ...................--------..((((.((((........((.(((((((((((((((((((........)))).))).)))))))))))))))))---------).))))... ( -25.00) >DroSec_CAF1 45617 103 + 1 GGAUAUAAAAAAAAAAAAC--------AGAAGGCGAAUAAAACAACGAUGGGGUUGUAAUGUCACUGUCAUUGCCAGCGGAUGAGUAAUGUAGCCCCUCAUU---------CGCCUUAUA ...................--------..(((((((((........((.(((((((((.(((((((((........)))).))).)).))))))))))))))---------))))))... ( -26.70) >DroSim_CAF1 46808 100 + 1 GGAUAUAAAAA---AAAAC--------AGAAGGCGAAUAAAACACCGAUGGGGUUGUAUUGUCACUGUCAUUGCCAGCGGAUGAGUAAUGUAGCCCCUCAUU---------CACCUUAUA ...........---.....--------..((((.((((........((.(((((((((((((((((((........)))).))).)))))))))))))))))---------).))))... ( -25.00) >DroEre_CAF1 47470 117 + 1 GGAUAUAAAAA---AAAAACAAAAAGAAAAAGGCGAAUAAAACACCAAUGGGGUUGUAUUGUCACUGUCAUUGCCAGCGGAUGAGUAAUGUAGACCCUCAUUUAGAAGAGACACCUUAUA ((.........---.................(((.((((.(((.((....))))).))))))).(((.((((((((.....)).))))))))).))(((........))).......... ( -18.50) >DroYak_CAF1 46281 109 + 1 GGAUAUAAAAA---AAAAC--------GGAAGUCGAAUAAAACACCGAUGGGGUUGUAUUGUCACUGUCAUUGCCAGCAGAUGAAUAAUGUAACCCCUCAUUUAGAAGAGCCACCUUAUA ((...((((..---...((--------....)).............((.(((((((((((((((((((........)))).)).))))))))))))))).))))......))........ ( -24.60) >consensus GGAUAUAAAAA___AAAAC________AGAAGGCGAAUAAAACACCGAUGGGGUUGUAUUGUCACUGUCAUUGCCAGCGGAUGAGUAAUGUAGCCCCUCAUU_________CACCUUAUA .............................((((.(..............(((((((((((((((((((........)))).))).))))))))))))..............).))))... (-19.35 = -19.63 + 0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:12:34 2006