
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,441,896 – 4,442,157 |
| Length | 261 |
| Max. P | 0.999843 |

| Location | 4,441,896 – 4,442,016 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.50 |
| Mean single sequence MFE | -31.14 |
| Consensus MFE | -25.39 |
| Energy contribution | -26.39 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.538351 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4441896 120 - 22224390 UCACGACAAUGGUGGUGAUCUCGUCGAGCUGCUUGUUUAUUUGCUGCCGUCGAGAUUUCAACGCCUACAUCUAUAGAUACUUAUGUGGCCGGAGUGCUAUUUAUAGACCUCACUCUCCAC ....((....((((..(((((((.((.((.((..........)).)))).)))))))....))))....)).............((((..(((((((((....)))....)))))))))) ( -32.80) >DroSec_CAF1 43542 120 - 1 UCACGACAAUGGUGGUGAUCUCGUCGAGCUGCUUGUUUAUUUGCUGCCGUCGAGAUUUCAACGCCUACAUCUAUAGAUACUUAUGUGGCCGGAGUGCUAUUUAUAGAUCUCACUCUCCAC ....((....((((..(((((((.((.((.((..........)).)))).)))))))....))))....)).............((((..((((((..(((....)))..)))))))))) ( -34.10) >DroSim_CAF1 44738 117 - 1 UCACGACAAUGGUGGUGAUCUCGUCGAGCUGCUUGUUUAUUUGCUGCC---GAGAUUUCAACGCCUACAUCUAUAGAUACUUAUGUGGCCGGAGUGCUAUUUAUAGAUCUCACUCUCCAC ....((....((((.(((..((.(((.((.((..........)).)))---))))..))).))))....)).............((((..((((((..(((....)))..)))))))))) ( -28.70) >DroEre_CAF1 45470 102 - 1 UCACGACAAUGGUGGUGAUCUCGUCGAGCUGCUUGUUUAUUUGCUGCCGUCGAGAUUUCAACGCCUACAUCUAUAG------------------UGCUAUUUAUAGACCCCACUCUCCUU ..........((((..(((((((.((.((.((..........)).)))).)))))))....))))....(((((((------------------......)))))))............. ( -27.50) >DroYak_CAF1 44270 119 - 1 UCACGACAAUGGUGGUGAUCUCGUCGAGCUGCUUGUUUAUUUGCUGCCGUCGAGAUUUCAACGCCUACAUCUAUAGAUACUUAUGCGGCCGGAGAGCUAUUUAUAGACCUCACUCUCC-U ...((.((..((((..(((((((.((.((.((..........)).)))).)))))))....))))...(((....))).....))))...((((((................))))))-. ( -32.59) >consensus UCACGACAAUGGUGGUGAUCUCGUCGAGCUGCUUGUUUAUUUGCUGCCGUCGAGAUUUCAACGCCUACAUCUAUAGAUACUUAUGUGGCCGGAGUGCUAUUUAUAGACCUCACUCUCCAC ..........((((..(((((((.((.((.((..........)).)))).)))))))....))))....((((((((((..................))))))))))............. (-25.39 = -26.39 + 1.00)

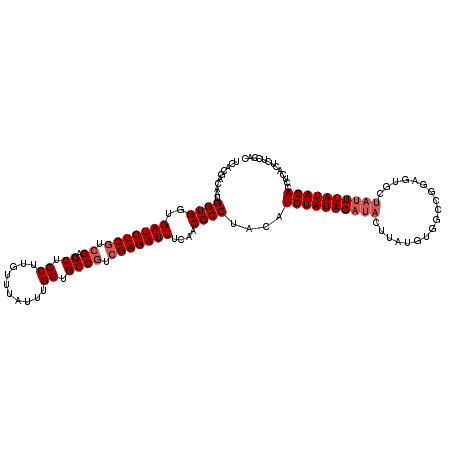
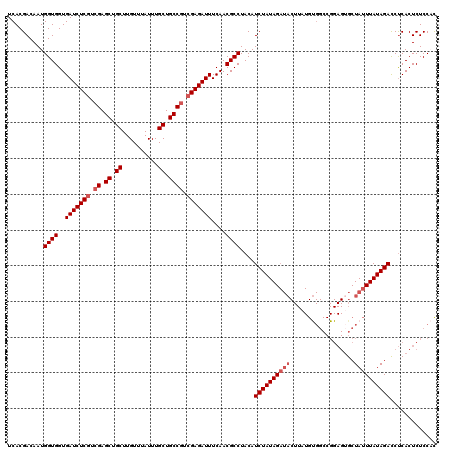
| Location | 4,441,936 – 4,442,056 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.00 |
| Mean single sequence MFE | -35.18 |
| Consensus MFE | -31.12 |
| Energy contribution | -32.92 |
| Covariance contribution | 1.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.25 |
| SVM RNA-class probability | 0.991176 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4441936 120 + 22224390 GUAUCUAUAGAUGUAGGCGUUGAAAUCUCGACGGCAGCAAAUAAACAAGCAGCUCGACGAGAUCACCACCAUUGUCGUGAUCUGCGCACAGCCACCGUUGAGACACAGAUACAUUCACAG ((((((.....(((.(.((((((....)))))).).)))............(((((((((((((((.((....)).)))))))((.....))...))))))).)..))))))........ ( -37.60) >DroSec_CAF1 43582 120 + 1 GUAUCUAUAGAUGUAGGCGUUGAAAUCUCGACGGCAGCAAAUAAACAAGCAGCUCGACGAGAUCACCACCAUUGUCGUGAUCUGCGCACAGCCACCGCUGAGGCACAGAUACAUUCACAG ((((((.....(((.(.((((((....)))))).).)))............(((.(.(((((((((.((....)).))))))).))).((((....)))).)))..))))))........ ( -35.90) >DroSim_CAF1 44778 117 + 1 GUAUCUAUAGAUGUAGGCGUUGAAAUCUC---GGCAGCAAAUAAACAAGCAGCUCGACGAGAUCACCACCAUUGUCGUGAUCUGCGCACAGCCACCGCUGAGGCACAGAUACAUUCACAG ((((((...((((....))))....((((---(((.((..........)).(((...(((((((((.((....)).))))))).))...)))....)))))))...))))))........ ( -32.20) >DroEre_CAF1 45496 116 + 1 ----CUAUAGAUGUAGGCGUUGAAAUCUCGACGGCAGCAAAUAAACAAGCAGCUCGACGAGAUCACCACCAUUGUCGUGAUCUGCGCACAGCCACCGUUGAGACAGAGAUACAUUCACAG ----.....(((((((.((((((....)))))).)................(((((((((((((((.((....)).)))))))((.....))...))))))).).....))))))..... ( -32.60) >DroYak_CAF1 44309 120 + 1 GUAUCUAUAGAUGUAGGCGUUGAAAUCUCGACGGCAGCAAAUAAACAAGCAGCUCGACGAGAUCACCACCAUUGUCGUGAUCUGCGCACAGCCACCGUUGAGACACAGAUACAUACACAG ((((((.....(((.(.((((((....)))))).).)))............(((((((((((((((.((....)).)))))))((.....))...))))))).)..))))))........ ( -37.60) >consensus GUAUCUAUAGAUGUAGGCGUUGAAAUCUCGACGGCAGCAAAUAAACAAGCAGCUCGACGAGAUCACCACCAUUGUCGUGAUCUGCGCACAGCCACCGUUGAGACACAGAUACAUUCACAG ((((((.....(((.(.((((((....)))))).).)))............(((((((((((((((.((....)).)))))))((.....))...))))))).)..))))))........ (-31.12 = -32.92 + 1.80)

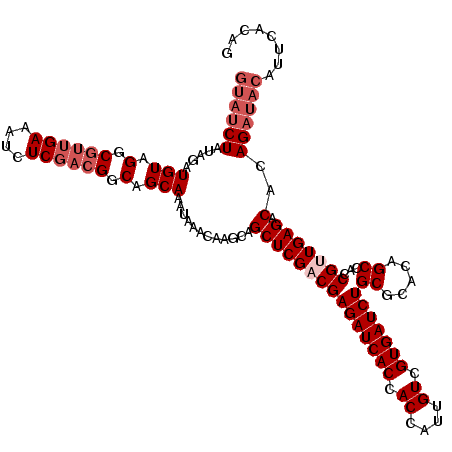
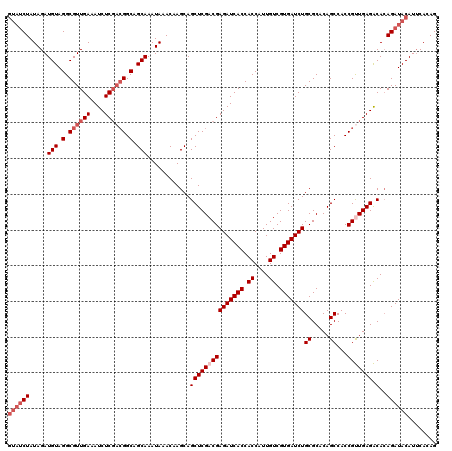
| Location | 4,441,936 – 4,442,056 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.00 |
| Mean single sequence MFE | -43.15 |
| Consensus MFE | -39.60 |
| Energy contribution | -40.48 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.02 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.92 |
| SVM decision value | 4.23 |
| SVM RNA-class probability | 0.999843 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4441936 120 - 22224390 CUGUGAAUGUAUCUGUGUCUCAACGGUGGCUGUGCGCAGAUCACGACAAUGGUGGUGAUCUCGUCGAGCUGCUUGUUUAUUUGCUGCCGUCGAGAUUUCAACGCCUACAUCUAUAGAUAC (((((.(((((..((((((((.((((..((...(((.(((((((.((....)).)))))))))).((((.....))))....))..)))).)))))....)))..))))).))))).... ( -43.80) >DroSec_CAF1 43582 120 - 1 CUGUGAAUGUAUCUGUGCCUCAGCGGUGGCUGUGCGCAGAUCACGACAAUGGUGGUGAUCUCGUCGAGCUGCUUGUUUAUUUGCUGCCGUCGAGAUUUCAACGCCUACAUCUAUAGAUAC (((((.((((((((((((..((((....)))).)))))))..........((((..(((((((.((.((.((..........)).)))).)))))))....))))))))).))))).... ( -46.60) >DroSim_CAF1 44778 117 - 1 CUGUGAAUGUAUCUGUGCCUCAGCGGUGGCUGUGCGCAGAUCACGACAAUGGUGGUGAUCUCGUCGAGCUGCUUGUUUAUUUGCUGCC---GAGAUUUCAACGCCUACAUCUAUAGAUAC (((((.((((((((((((..((((....)))).)))))))..........((((.(((..((.(((.((.((..........)).)))---))))..))).))))))))).))))).... ( -41.20) >DroEre_CAF1 45496 116 - 1 CUGUGAAUGUAUCUCUGUCUCAACGGUGGCUGUGCGCAGAUCACGACAAUGGUGGUGAUCUCGUCGAGCUGCUUGUUUAUUUGCUGCCGUCGAGAUUUCAACGCCUACAUCUAUAG---- (((((.(((((.....(((((.((((..((...(((.(((((((.((....)).)))))))))).((((.....))))....))..)))).))))).........))))).)))))---- ( -40.34) >DroYak_CAF1 44309 120 - 1 CUGUGUAUGUAUCUGUGUCUCAACGGUGGCUGUGCGCAGAUCACGACAAUGGUGGUGAUCUCGUCGAGCUGCUUGUUUAUUUGCUGCCGUCGAGAUUUCAACGCCUACAUCUAUAGAUAC (((((.(((((..((((((((.((((..((...(((.(((((((.((....)).)))))))))).((((.....))))....))..)))).)))))....)))..))))).))))).... ( -43.80) >consensus CUGUGAAUGUAUCUGUGUCUCAACGGUGGCUGUGCGCAGAUCACGACAAUGGUGGUGAUCUCGUCGAGCUGCUUGUUUAUUUGCUGCCGUCGAGAUUUCAACGCCUACAUCUAUAGAUAC (((((.(((((..((((((((.((((..((...(((.(((((((.((....)).)))))))))).((((.....))))....))..)))).)))))....)))..))))).))))).... (-39.60 = -40.48 + 0.88)


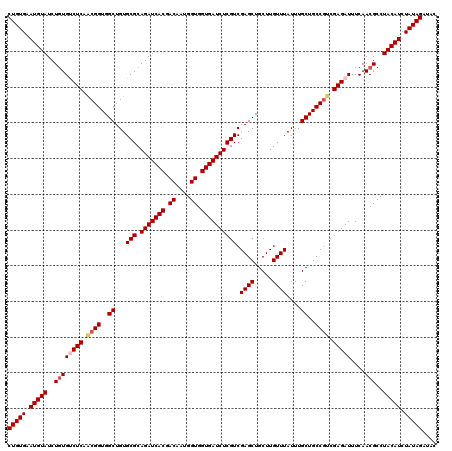
| Location | 4,441,976 – 4,442,091 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 95.40 |
| Mean single sequence MFE | -26.89 |
| Consensus MFE | -23.14 |
| Energy contribution | -23.25 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.95 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.909393 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4441976 115 + 22224390 AUAAACAAGCAGCUCGACGAGAUCACCACCAUUGUCGUGAUCUGCGCACAGCCACCGUUGAGACACAGAUACAUUCACAGAUACAGCAAAAAAAUAAAU--AAAAAGUAUCUGAAAG ...........(((((((((((((((.((....)).)))))))((.....))...))))))).).............(((((((...............--.....))))))).... ( -27.15) >DroSec_CAF1 43622 116 + 1 AUAAACAAGCAGCUCGACGAGAUCACCACCAUUGUCGUGAUCUGCGCACAGCCACCGCUGAGGCACAGAUACAUUCACAGAUACAGCUAAAAA-CAAAACAAAAAAGUAUCUGAAAG ............(.((((((...........)))))).)(((((.((.((((....))))..)).))))).......(((((((.........-............))))))).... ( -26.70) >DroSim_CAF1 44815 114 + 1 AUAAACAAGCAGCUCGACGAGAUCACCACCAUUGUCGUGAUCUGCGCACAGCCACCGCUGAGGCACAGAUACAUUCACAGAUACAGCUAAAAA-UAAAA--AAAAAGUAUCUGAAAG ............(.((((((...........)))))).)(((((.((.((((....))))..)).))))).......(((((((.........-.....--.....))))))).... ( -26.81) >consensus AUAAACAAGCAGCUCGACGAGAUCACCACCAUUGUCGUGAUCUGCGCACAGCCACCGCUGAGGCACAGAUACAUUCACAGAUACAGCUAAAAA_UAAAA__AAAAAGUAUCUGAAAG ............(.((((((...........)))))).)(((((.((.((((....))))..)).))))).......(((((((......................))))))).... (-23.14 = -23.25 + 0.11)

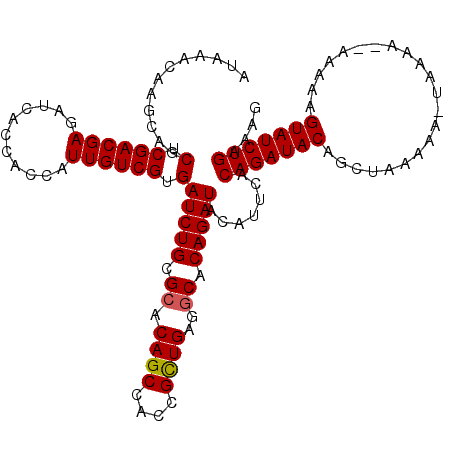

| Location | 4,441,976 – 4,442,091 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 95.40 |
| Mean single sequence MFE | -33.13 |
| Consensus MFE | -31.83 |
| Energy contribution | -31.38 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.921299 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4441976 115 - 22224390 CUUUCAGAUACUUUUU--AUUUAUUUUUUUGCUGUAUCUGUGAAUGUAUCUGUGUCUCAACGGUGGCUGUGCGCAGAUCACGACAAUGGUGGUGAUCUCGUCGAGCUGCUUGUUUAU ....(((((((..(((--((.(((.........)))...))))).)))))))......(((((..(((..(((.(((((((.((....)).))))))))))..)))..).))))... ( -33.90) >DroSec_CAF1 43622 116 - 1 CUUUCAGAUACUUUUUUGUUUUG-UUUUUAGCUGUAUCUGUGAAUGUAUCUGUGCCUCAGCGGUGGCUGUGCGCAGAUCACGACAAUGGUGGUGAUCUCGUCGAGCUGCUUGUUUAU ....(((((((......(((...-.....))).)))))))(((((....(((.....))).((..(((..(((.(((((((.((....)).))))))))))..)))..)).))))). ( -32.50) >DroSim_CAF1 44815 114 - 1 CUUUCAGAUACUUUUU--UUUUA-UUUUUAGCUGUAUCUGUGAAUGUAUCUGUGCCUCAGCGGUGGCUGUGCGCAGAUCACGACAAUGGUGGUGAUCUCGUCGAGCUGCUUGUUUAU ....(((((((.(((.--...((-(........))).....))).)))))))......(((((..(((..(((.(((((((.((....)).))))))))))..)))..).))))... ( -33.00) >consensus CUUUCAGAUACUUUUU__UUUUA_UUUUUAGCUGUAUCUGUGAAUGUAUCUGUGCCUCAGCGGUGGCUGUGCGCAGAUCACGACAAUGGUGGUGAUCUCGUCGAGCUGCUUGUUUAU ....(((((((......................)))))))(((((........((....))((..(((..(((.(((((((.((....)).))))))))))..)))..)).))))). (-31.83 = -31.38 + -0.44)


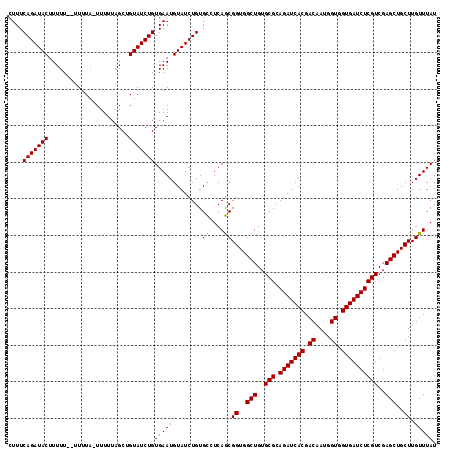
| Location | 4,442,016 – 4,442,128 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 91.95 |
| Mean single sequence MFE | -27.35 |
| Consensus MFE | -24.69 |
| Energy contribution | -24.58 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.671630 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4442016 112 + 22224390 UCUGCGCACAGCCACCGUUGAGACACAGAUACAUUCACAGAUACAGCAAAAAAAUAAAU--AAAAAGUAUCUGAAAGAUACGAGUUCGUCUGCGGUCGGGCUCCCCAUGUACAU--- ((((.((.((((....)))).).).))))(((((...(((((((...............--.....)))))))........(((((((.(....).)))))))...)))))...--- ( -25.55) >DroSec_CAF1 43662 116 + 1 UCUGCGCACAGCCACCGCUGAGGCACAGAUACAUUCACAGAUACAGCUAAAAA-CAAAACAAAAAAGUAUCUGAAAGAUACGAGUUCGUCUGCGGUCGGGCUCCCCAUAUACAUACA ((((.((.((((....))))..)).))))........(((((((.........-............)))))))........(((((((.(....).))))))).............. ( -28.70) >DroSim_CAF1 44855 114 + 1 UCUGCGCACAGCCACCGCUGAGGCACAGAUACAUUCACAGAUACAGCUAAAAA-UAAAA--AAAAAGUAUCUGAAAGAUACGAGUUUGUCUGCGGUCGGGCUCCCCAUGCACAUACA ..((.(((((((....)))).(((.((((((.((((..((......)).....-.....--.....(((((.....))))))))).))))))..)))(((...))).))).)).... ( -27.80) >consensus UCUGCGCACAGCCACCGCUGAGGCACAGAUACAUUCACAGAUACAGCUAAAAA_UAAAA__AAAAAGUAUCUGAAAGAUACGAGUUCGUCUGCGGUCGGGCUCCCCAUGUACAUACA ((((.((.((((....))))..)).))))........(((((((......................)))))))........(((((((.(....).))))))).............. (-24.69 = -24.58 + -0.11)

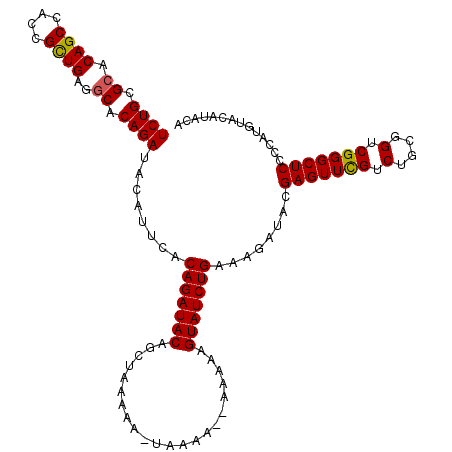
| Location | 4,442,056 – 4,442,157 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 86.49 |
| Mean single sequence MFE | -27.10 |
| Consensus MFE | -22.04 |
| Energy contribution | -24.27 |
| Covariance contribution | 2.23 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.914213 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4442056 101 + 22224390 AUACAGCAAAAAAAUAAAU--AAAAAGUAUCUGAAAGAUACGAGUUCGUCUGCGGUCGGGCUCCCCAUGUACAU--------------AUAUGUAUAAUAGUGUAGCAUGGUAUGGU ...................--.....(((((.....)))))(((((((.(....).))))))).((((((((((--------------.(((.....))))))).))))))...... ( -24.30) >DroSec_CAF1 43702 116 + 1 AUACAGCUAAAAA-CAAAACAAAAAAGUAUCUGAAAGAUACGAGUUCGUCUGCGGUCGGGCUCCCCAUAUACAUACAUACAUACAUAGAUAUGUAUAAUAGUGUAGCAUGGUAUGGU ((((.((((...(-(...........(((((.....)))))(((((((.(....).)))))))....((((((((..((......))..))))))))...)).))))...))))... ( -27.30) >DroSim_CAF1 44895 114 + 1 AUACAGCUAAAAA-UAAAA--AAAAAGUAUCUGAAAGAUACGAGUUUGUCUGCGGUCGGGCUCCCCAUGCACAUACAUACAUACAUAUAUAUGUAUAAUAGUGUAGCAUGGUAUGGU .....((((....-.....--.....(((((.....)))))(((((((.(....).))))))).((((((((((..(((((((......)))))))....)))).))))))..)))) ( -29.70) >consensus AUACAGCUAAAAA_UAAAA__AAAAAGUAUCUGAAAGAUACGAGUUCGUCUGCGGUCGGGCUCCCCAUGUACAUACAUACAUACAUA_AUAUGUAUAAUAGUGUAGCAUGGUAUGGU ..........................(((((.....)))))(((((((.(....).))))))).((((((((((..(((((((......)))))))....)))).))))))...... (-22.04 = -24.27 + 2.23)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:12:30 2006