
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,441,555 – 4,441,742 |
| Length | 187 |
| Max. P | 0.977952 |

| Location | 4,441,555 – 4,441,662 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.00 |
| Mean single sequence MFE | -36.24 |
| Consensus MFE | -29.96 |
| Energy contribution | -30.36 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.959988 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4441555 107 + 22224390 AAAACUUUGGACCCAUGCAUCGUCGACCGGUCCAAGACAGGGGCGUGGGCGAAA-GA------------GGGGGAUCCAAAGCAGAGUGGCGUGUGAAAAAUGCCAAGUGGCUUUAAAUG ........((((((.....(((((.((.(.(((......))).))).)))))..-..------------..))).)))(((((....((((((.......))))))....)))))..... ( -36.30) >DroSec_CAF1 43174 119 + 1 AAAACUUUGGGCCCAUGCAUCGUCGACUGGUCCAAGACAGGGGCGUGGGCGAAA-GAUGGGGGGGGGGGGGGGGAUCCAAAGCAGAGUGGCGUGUGAAAAAUGCCAAGUGGCUUUAAAUG ....((((...(((((...(((((.((.(.(((......))).))).)))))..-.)))))...))))(((....)))(((((....((((((.......))))))....)))))..... ( -37.00) >DroSim_CAF1 44363 113 + 1 AAAACUUUGGGCCCAUGCAUCGUCGACUGGUCCAAGACAGGGGCGUGGGCGAAA-GACGGGGG------GGGGGAUCCAAAGCAGAGUGGCGUGUGAAAAAUGCCAAGUGGCUUUAAAUG ....((((.(.(((((((..((((...........))).)..)))))))).)))-)..(((..------......)))(((((....((((((.......))))))....)))))..... ( -36.60) >DroEre_CAF1 45053 108 + 1 AAAACUUUGGGCCCAUGCAUCGUCGACUGGACCAAGACAGGGGCGUGGGCGAACAUA------------GGGGGACCUAAAGCAGUGUGGCGUGUGAAAAAUGCCAAGUGGCUUUAAAUG ..........((((((((..((((...........))).)..)))))))).......------------((....))((((((....((((((.......))))))....)))))).... ( -39.20) >DroYak_CAF1 43914 107 + 1 AAAACUUUGGACCCAUGCAUCGUCGACUGGACCAAGACAGGGGCGUGGGCGAAA-AA------------GGGGGACCCUAAGCAGAGUGGCGUGUGAAAAAUGCCAAGUGACUUUAAAUG ...(((((((.(((((((..((((...........))).)..))))))))....-..------------((....)).....))))))(((((.......)))))............... ( -32.10) >consensus AAAACUUUGGGCCCAUGCAUCGUCGACUGGUCCAAGACAGGGGCGUGGGCGAAA_GA____________GGGGGAUCCAAAGCAGAGUGGCGUGUGAAAAAUGCCAAGUGGCUUUAAAUG ..........((((((((..((((...........))).)..))))))))...................(((...)))(((((....((((((.......))))))....)))))..... (-29.96 = -30.36 + 0.40)

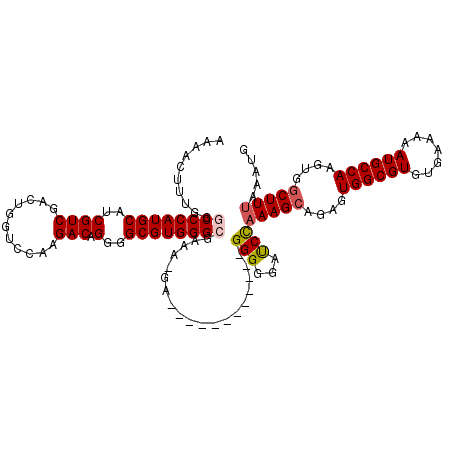

| Location | 4,441,595 – 4,441,702 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.33 |
| Mean single sequence MFE | -24.53 |
| Consensus MFE | -20.92 |
| Energy contribution | -21.76 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.977952 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4441595 107 - 22224390 CAAAGAAAAUCAAUUUUGCAGAAGCAGCAGUGGCAGCACACAUUUAAAGCCACUUGGCAUUUUUCACACGCCACUCUGCUUUGGAUCCCCC------------UC-UUUCGCCCACGCCC .(((((..............(((((((.((((((.(....).......(((....)))...........)))))))))))))((.....))------------))-)))........... ( -26.10) >DroSec_CAF1 43214 119 - 1 CAAAGAAAAUCAAUUUUGCAGAAGCAGCAGUGGCAGCACACAUUUAAAGCCACUUGGCAUUUUUCACACGCCACUCUGCUUUGGAUCCCCCCCCCCCCCCCCAUC-UUUCGCCCACGCCC .(((((..............(((((((.((((((.(....).......(((....)))...........)))))))))))))((......))...........))-)))........... ( -26.10) >DroSim_CAF1 44403 113 - 1 CAAAGAAAAUCAAUUUUGCAGAAGCAGCAGUGGCAGCACACAUUUAAAGCCACUUGGCAUUUUUCACACGCCACUCUGCUUUGGAUCCCCC------CCCCCGUC-UUUCGCCCACGCCC ....((((..(.........(((((((.((((((.(....).......(((....)))...........)))))))))))))((......)------)....)..-)))).......... ( -26.40) >DroEre_CAF1 45093 106 - 1 CAAAGAAAAUCAAUUUUGCAGAAGCA--AGUGGCAGCACACAUUUAAAGCCACUUGGCAUUUUUCACACGCCACACUGCUUUAGGUCCCCC------------UAUGUUCGCCCACGCCC ....(((..........((((....(--((((((..............)))))))(((...........)))...))))..((((....))------------))..))).......... ( -21.74) >DroYak_CAF1 43954 105 - 1 CAAAGAAAAUCAAUUUUGCAGAAGCA--AGUGGCAGCACACAUUUAAAGUCACUUGGCAUUUUUCACACGCCACUCUGCUUAGGGUCCCCC------------UU-UUUCGCCCACGCCC .................((..(((((--((((((.(....).......(((....)))...........)))))).))))).((((.....------------..-....))))..)).. ( -22.30) >consensus CAAAGAAAAUCAAUUUUGCAGAAGCAGCAGUGGCAGCACACAUUUAAAGCCACUUGGCAUUUUUCACACGCCACUCUGCUUUGGAUCCCCC____________UC_UUUCGCCCACGCCC .................((.(((((((.((((((.(....).......(((....)))...........)))))))))))))((....))....................))........ (-20.92 = -21.76 + 0.84)

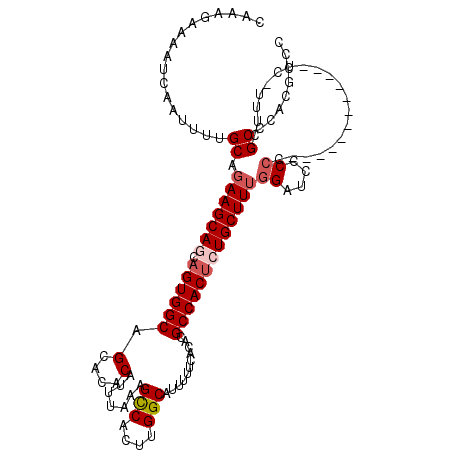

| Location | 4,441,622 – 4,441,742 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.99 |
| Mean single sequence MFE | -28.88 |
| Consensus MFE | -24.84 |
| Energy contribution | -25.32 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.712381 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4441622 120 - 22224390 GCUGAGUUUUCGGAUGGAGUUACAUAAAUAUUCGACGAGACAAAGAAAAUCAAUUUUGCAGAAGCAGCAGUGGCAGCACACAUUUAAAGCCACUUGGCAUUUUUCACACGCCACUCUGCU .(((.(((((((((((............)))))))..))))...(.....).......))).(((((.((((((.(....).......(((....)))...........))))))))))) ( -27.50) >DroSec_CAF1 43253 120 - 1 CCUGAGUUUUCGGAUGGAGUUACAUAAAUAUCCGACAAGACAAAGAAAAUCAAUUUUGCAGAAGCAGCAGUGGCAGCACACAUUUAAAGCCACUUGGCAUUUUUCACACGCCACUCUGCU .(((.(((((((((((............)))))))..))))...(.....).......))).(((((.((((((.(....).......(((....)))...........))))))))))) ( -30.50) >DroSim_CAF1 44436 120 - 1 CCUGAGUUUUCGGAUGGAGUUACAUAAAUAUCCGACAAGACAAAGAAAAUCAAUUUUGCAGAAGCAGCAGUGGCAGCACACAUUUAAAGCCACUUGGCAUUUUUCACACGCCACUCUGCU .(((.(((((((((((............)))))))..))))...(.....).......))).(((((.((((((.(....).......(((....)))...........))))))))))) ( -30.50) >DroEre_CAF1 45121 118 - 1 GCAGAGUUUUCGGAUGGAGUUACAUAGAUAUCCGACAAGACAAAGAAAAUCAAUUUUGCAGAAGCA--AGUGGCAGCACACAUUUAAAGCCACUUGGCAUUUUUCACACGCCACACUGCU ((((.(((((((((((............)))))))..))))..............((((....)))--)(((((.(....).......(((....)))...........))))).)))). ( -29.70) >DroYak_CAF1 43981 118 - 1 GCAGAGUUUUCGGAUGGAGUUACAUAAAUAUCGGACAAGACAAAGAAAAUCAAUUUUGCAGAAGCA--AGUGGCAGCACACAUUUAAAGUCACUUGGCAUUUUUCACACGCCACUCUGCU (((((((..((.((((............)))).))...(((..............((((....)))--)(((......))).......)))....(((...........)))))))))). ( -26.20) >consensus GCUGAGUUUUCGGAUGGAGUUACAUAAAUAUCCGACAAGACAAAGAAAAUCAAUUUUGCAGAAGCAGCAGUGGCAGCACACAUUUAAAGCCACUUGGCAUUUUUCACACGCCACUCUGCU .....(((((((((((............)))))))..)))).....................(((((.((((((.(....).......(((....)))...........))))))))))) (-24.84 = -25.32 + 0.48)

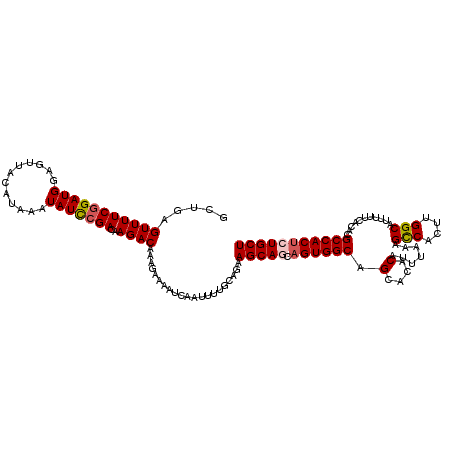

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:12:23 2006