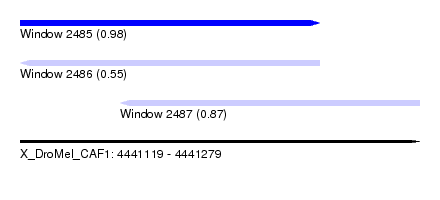
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,441,119 – 4,441,279 |
| Length | 160 |
| Max. P | 0.981172 |

| Location | 4,441,119 – 4,441,239 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.33 |
| Mean single sequence MFE | -24.30 |
| Consensus MFE | -19.44 |
| Energy contribution | -19.52 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.88 |
| SVM RNA-class probability | 0.981172 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4441119 120 + 22224390 CUAAUUUAAUUUUCUGAUCACUUGAGUGAAUAUAUCUUAGUGCCCGUAUUCACCAUAAAUCUUCUCGACAAAGCAAACACCCGCUUGUGAUCCACGAAAACUGCUAUACCCAAGCAGAUU .........(((((.((((((.((.((((((((............))))))))))...............((((........))))))))))...)))))(((((.......)))))... ( -25.10) >DroSec_CAF1 42736 120 + 1 UUAAUUUUAUUUUCUGAUCACUUGAGUGAAUACAUUUUAGAGCCCGUAUUCGCCAUAAAUCUUCUCGACAAAACAAACACCCGCUUGUGAUCCACGAAAACUGCUAUACCCAAGCAGAUU .........(((((.((((((.((.((((((((............))))))))))....((.....))..................))))))...)))))(((((.......)))))... ( -22.40) >DroSim_CAF1 43925 120 + 1 AUAAUUUAAUUUUCUGAUCACUUGAGUGAAUACAUUUUACAGCCCGUAUUCGCCAUAAAUCUUCUCGGCAAAACAAACACCCGCUUGUGAUCCACGAAAACUGCUAUACCCAAGCAGAUU .........(((((.((((((..((((((((((............))))))(((............))).............))))))))))...)))))(((((.......)))))... ( -25.20) >DroEre_CAF1 44617 119 + 1 CUAAUUUAAU-UUCUGAUCACUUGAGGGAAUACCUUUUAGUGGCUGUAUUCGCCAUAAAUCUUUUCGACAAAACAAACACCCGCUUGUGAUCCACGAAAACUGCUAUACCUUAGCAGAUU .........(-(((.((((((..(.(((...........(((((.......)))))...((.....))...........))).)..))))))...)))).((((((.....))))))... ( -29.20) >DroYak_CAF1 43483 118 + 1 CUAAUUUCAUUUUCUGAUCACUUGAGUGAAUAUAUUUUCGUGCCUGUAUUCGCCAUAAAUCUUCUCGACAAAAUAAACACCCGCUUGUGAUCCACGAAAACUGCUAUACCUUCGCAG--U ..........((((.((((((.((.(((((((((..........)))))))))))....((.....))..................))))))...))))(((((.........))))--) ( -19.60) >consensus CUAAUUUAAUUUUCUGAUCACUUGAGUGAAUACAUUUUAGUGCCCGUAUUCGCCAUAAAUCUUCUCGACAAAACAAACACCCGCUUGUGAUCCACGAAAACUGCUAUACCCAAGCAGAUU .........(((((.((((((.((.((((((((............))))))))))....((.....))..................))))))...)))))(((((.......)))))... (-19.44 = -19.52 + 0.08)

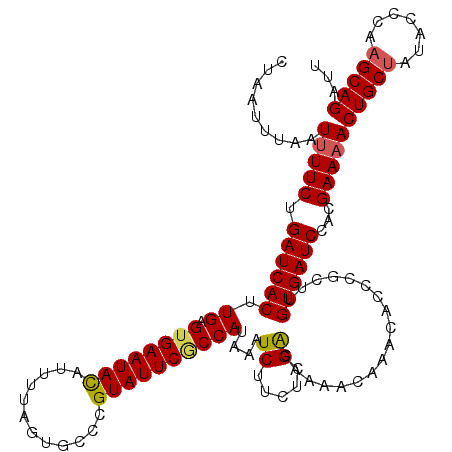

| Location | 4,441,119 – 4,441,239 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.33 |
| Mean single sequence MFE | -29.88 |
| Consensus MFE | -23.42 |
| Energy contribution | -24.22 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.545914 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4441119 120 - 22224390 AAUCUGCUUGGGUAUAGCAGUUUUCGUGGAUCACAAGCGGGUGUUUGCUUUGUCGAGAAGAUUUAUGGUGAAUACGGGCACUAAGAUAUAUUCACUCAAGUGAUCAGAAAAUUAAAUUAG ...(((((.......)))))(((((...((((((..((..(((((..((..(((.....)))....))..)))))..)).....((.....))......)))))).)))))......... ( -30.90) >DroSec_CAF1 42736 120 - 1 AAUCUGCUUGGGUAUAGCAGUUUUCGUGGAUCACAAGCGGGUGUUUGUUUUGUCGAGAAGAUUUAUGGCGAAUACGGGCUCUAAAAUGUAUUCACUCAAGUGAUCAGAAAAUAAAAUUAA ...(((((.......)))))(((((...((((((.(((..(((((((((..(((.....)))....)))))))))..)))......((........)).)))))).)))))......... ( -29.50) >DroSim_CAF1 43925 120 - 1 AAUCUGCUUGGGUAUAGCAGUUUUCGUGGAUCACAAGCGGGUGUUUGUUUUGCCGAGAAGAUUUAUGGCGAAUACGGGCUGUAAAAUGUAUUCACUCAAGUGAUCAGAAAAUUAAAUUAU ..(((((((((((...(((((((((.(((...((((((....))))))....))).)))))))....))(((((((..........))))))))))))))....))))............ ( -29.60) >DroEre_CAF1 44617 119 - 1 AAUCUGCUAAGGUAUAGCAGUUUUCGUGGAUCACAAGCGGGUGUUUGUUUUGUCGAAAAGAUUUAUGGCGAAUACAGCCACUAAAAGGUAUUCCCUCAAGUGAUCAGAA-AUUAAAUUAG ...((((((.....)))))).((((...((((((..(.(((((((((((..(((.....)))....))))))))).(((.......)))....)).)..)))))).)))-)......... ( -31.80) >DroYak_CAF1 43483 118 - 1 A--CUGCGAAGGUAUAGCAGUUUUCGUGGAUCACAAGCGGGUGUUUAUUUUGUCGAGAAGAUUUAUGGCGAAUACAGGCACGAAAAUAUAUUCACUCAAGUGAUCAGAAAAUGAAAUUAG (--((((.........)))))((((((.((((((......((((((..(((((((.((....)).)))))))...))))))(((......)))......)))))).....)))))).... ( -27.60) >consensus AAUCUGCUUGGGUAUAGCAGUUUUCGUGGAUCACAAGCGGGUGUUUGUUUUGUCGAGAAGAUUUAUGGCGAAUACGGGCACUAAAAUGUAUUCACUCAAGUGAUCAGAAAAUUAAAUUAG ...(((((.......)))))(((((...((((((..((..(((((((((..(((.....)))....)))))))))..)).......((....)).....)))))).)))))......... (-23.42 = -24.22 + 0.80)


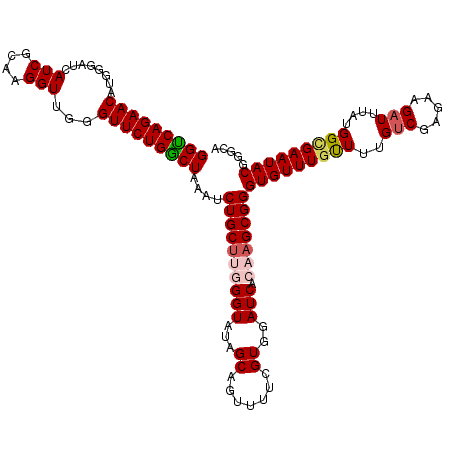
| Location | 4,441,159 – 4,441,279 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.00 |
| Mean single sequence MFE | -36.82 |
| Consensus MFE | -31.62 |
| Energy contribution | -32.42 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.872737 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4441159 120 - 22224390 GGUCAGAACAUGGGAUCAUCGCAAGGUUGGGUUCUGACUAAAUCUGCUUGGGUAUAGCAGUUUUCGUGGAUCACAAGCGGGUGUUUGCUUUGUCGAGAAGAUUUAUGGUGAAUACGGGCA (((((((((........(((....)))...)))))))))....((((((((((...((.......))..))).)))))))(((((..((..(((.....)))....))..)))))..... ( -37.40) >DroSec_CAF1 42776 120 - 1 GGUCAGAACAUGGGAUCAUCGCAAGGUUGGGUUCUGGCUAAAUCUGCUUGGGUAUAGCAGUUUUCGUGGAUCACAAGCGGGUGUUUGUUUUGUCGAGAAGAUUUAUGGCGAAUACGGGCU (((((((((........(((....)))...)))))))))..((((((((((((...((.......))..))).)))))))))(((((((((((((.((....)).))))))).)))))). ( -36.90) >DroSim_CAF1 43965 120 - 1 GGUCAGAACAUGGGAUCAUCGCAAGGUUGGGUUCUGGCUAAAUCUGCUUGGGUAUAGCAGUUUUCGUGGAUCACAAGCGGGUGUUUGUUUUGCCGAGAAGAUUUAUGGCGAAUACGGGCU (((((((((........(((....)))...)))))))))..((((((((((((...((.......))..))).)))))))))(((((((((((((.((....)).))))))).)))))). ( -39.60) >DroEre_CAF1 44656 120 - 1 GGCCAGAACAUGGGAUCAUCGCAAGGUUGUGUUCUGGCUAAAUCUGCUAAGGUAUAGCAGUUUUCGUGGAUCACAAGCGGGUGUUUGUUUUGUCGAAAAGAUUUAUGGCGAAUACAGCCA ((((((((((((.....(((....)))))))))))))))((((((((((.....))))).((((((......((((((....)))))).....))))))))))).((((.......)))) ( -39.50) >DroYak_CAF1 43523 118 - 1 GGUCAGAACAUGGGAUCAUCGCAAGGUUGGGUUCUGGCUAA--CUGCGAAGGUAUAGCAGUUUUCGUGGAUCACAAGCGGGUGUUUAUUUUGUCGAGAAGAUUUAUGGCGAAUACAGGCA (((((((((........(((....)))...)))))))))..--(..((((..(......)..))))..).......((..(((((..((..(((.....)))....))..)))))..)). ( -30.70) >consensus GGUCAGAACAUGGGAUCAUCGCAAGGUUGGGUUCUGGCUAAAUCUGCUUGGGUAUAGCAGUUUUCGUGGAUCACAAGCGGGUGUUUGUUUUGUCGAGAAGAUUUAUGGCGAAUACGGGCA (((((((((........(((....)))...)))))))))....((((((((((...((.......))..))).)))))))(((((((((..(((.....)))....)))))))))..... (-31.62 = -32.42 + 0.80)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:12:21 2006