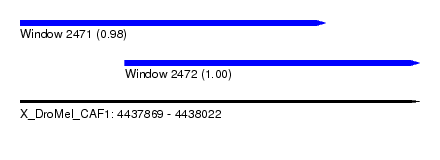
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,437,869 – 4,438,022 |
| Length | 153 |
| Max. P | 0.996327 |

| Location | 4,437,869 – 4,437,986 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.45 |
| Mean single sequence MFE | -24.63 |
| Consensus MFE | -24.50 |
| Energy contribution | -24.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.977626 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4437869 117 + 22224390 GGCAUGUGCAAAAUAUUUCACGGCUUGACACCUUCUUCGGUUUCUGGUUACGCUCACAGCCUCAUGGCGCCACUCGUAAUUAGAACGCCCCAAUAAU---AAUAAUCCCCAUCACACAUU ((...(((.((....)).)))(((.....(((......)))(((((((((((......(((....)))......))))))))))).)))))......---.................... ( -24.50) >DroSec_CAF1 39680 117 + 1 GGCAUGUGCAAAAUAUUUCACGGCUUGACACCUUCUUCGGUUUCUGGUUACGCUCACAGCCUCAUGGCGCCACUCGUAAUUAGAACGCCCCAAUAAU---CAUAAUCCCCAUCCCACACU ((...(((.((....)).)))(((.....(((......)))(((((((((((......(((....)))......))))))))))).)))))......---.................... ( -24.50) >DroSim_CAF1 41112 117 + 1 GGCAUGUGCAAAAUAUUUCACGGCUUGACACCUUAUUCGGUUUCUGGUUACGCUCACAGCCUCAUGGCGCCACUCGUAAUUAGAACGCCCCAAUAAU---AAUAAUCCCCAUCCCACACU ((.(((.......((((....(((.....(((......)))(((((((((((......(((....)))......))))))))))).))).....)))---)........))).))..... ( -25.16) >DroEre_CAF1 41292 117 + 1 GGCAUGUGCAAAAUAUUUCACGGCUUGACACCUUCUUCGGUUUCUGGUUACGCUCACAGCCUCAUGGCGCCACUCGUAAUUAGAACGCCCCAAUAAU---AAUAAUCCCCAUCUCAAACU ((...(((.((....)).)))(((.....(((......)))(((((((((((......(((....)))......))))))))))).)))))......---.................... ( -24.50) >DroYak_CAF1 40302 120 + 1 GGCAUGUGCAAAAUAUUUCACGGCUUGACACCUUCUUCGGUUUCUGGUUACGCUCACAGCCUCAUGGCGCCACUCGUAAUUAGAACGCCCCAAUAAUAAUAAUAAUCCCCAUCCUAUACU ((...(((.((....)).)))(((.....(((......)))(((((((((((......(((....)))......))))))))))).)))))............................. ( -24.50) >consensus GGCAUGUGCAAAAUAUUUCACGGCUUGACACCUUCUUCGGUUUCUGGUUACGCUCACAGCCUCAUGGCGCCACUCGUAAUUAGAACGCCCCAAUAAU___AAUAAUCCCCAUCCCACACU ((...(((.((....)).)))(((.....(((......)))(((((((((((......(((....)))......))))))))))).)))))............................. (-24.50 = -24.50 + 0.00)


| Location | 4,437,909 – 4,438,022 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.19 |
| Mean single sequence MFE | -17.98 |
| Consensus MFE | -17.30 |
| Energy contribution | -17.30 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.68 |
| SVM RNA-class probability | 0.996327 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4437909 113 + 22224390 UUUCUGGUUACGCUCACAGCCUCAUGGCGCCACUCGUAAUUAGAACGCCCCAAUAAU---AAUAAUCCCCAUCACACAUUCCCAUCCCCUCCACUUCAUUUCUCUG----UCUGGCUUUG .(((((((((((......(((....)))......))))))))))).(((........---..............................................----...))).... ( -17.42) >DroSec_CAF1 39720 114 + 1 UUUCUGGUUACGCUCACAGCCUCAUGGCGCCACUCGUAAUUAGAACGCCCCAAUAAU---CAUAAUCCCCAUCCCACACUCCCAUCCC---CACUUCAAUUCUCUGUCUGUCUGGCUUUG .(((((((((((......(((....)))......)))))))))))............---............................---..............(((.....))).... ( -18.40) >DroSim_CAF1 41152 114 + 1 UUUCUGGUUACGCUCACAGCCUCAUGGCGCCACUCGUAAUUAGAACGCCCCAAUAAU---AAUAAUCCCCAUCCCACACUCCCAUCCC---CACUUCAUUUCUCUGUCUGUCUGGCUUUG .(((((((((((......(((....)))......)))))))))))............---............................---..............(((.....))).... ( -18.40) >DroEre_CAF1 41332 110 + 1 UUUCUGGUUACGCUCACAGCCUCAUGGCGCCACUCGUAAUUAGAACGCCCCAAUAAU---AAUAAUCCCCAUCUCAAACUCCCAUCCC---CACUUCAUUUCUCUG----UCUGGCUUUG .(((((((((((......(((....)))......)))))))))))............---...........................(---((...((......))----..)))..... ( -17.60) >DroYak_CAF1 40342 113 + 1 UUUCUGGUUACGCUCACAGCCUCAUGGCGCCACUCGUAAUUAGAACGCCCCAAUAAUAAUAAUAAUCCCCAUCCUAUACUCCCAUCCC---CACUGCUUUUUUCUG----UCUGGCUUUG .(((((((((((......(((....)))......)))))))))))..........................................(---((..((........)----).)))..... ( -18.10) >consensus UUUCUGGUUACGCUCACAGCCUCAUGGCGCCACUCGUAAUUAGAACGCCCCAAUAAU___AAUAAUCCCCAUCCCACACUCCCAUCCC___CACUUCAUUUCUCUG____UCUGGCUUUG .(((((((((((......(((....)))......))))))))))).(((................................................................))).... (-17.30 = -17.30 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:12:07 2006