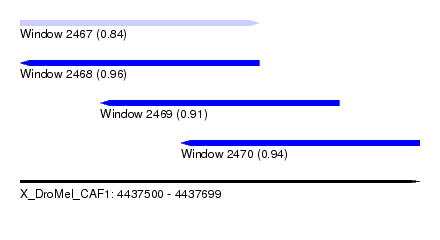
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,437,500 – 4,437,699 |
| Length | 199 |
| Max. P | 0.955501 |

| Location | 4,437,500 – 4,437,619 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.43 |
| Mean single sequence MFE | -25.26 |
| Consensus MFE | -18.98 |
| Energy contribution | -21.04 |
| Covariance contribution | 2.06 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.835746 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4437500 119 + 22224390 GUUUUCAUUUCUUCUUUUUUUUUUUAGCCCCUUCCCCCAAUGGCAUGCUGCAUGUCUUCAUUAUCGUUUAUUGAAUACAAGCAUGCUUAUACACAUGUAUGUGUGCGUGUAGAGAGA-AU ...........((((((((......................((((((((...(((.((((.((.....)).)))).))))))))))).....(((((((....))))))))))))))-). ( -26.20) >DroSec_CAF1 39332 115 + 1 GUUUUCAUUUCU-----UUUUUUUUAGCCCCUUCCCCCAAUGGCAUGCUGCAUGUCUUCAUUAUCGUUUAUUAAAUACAAGCAUGCUUAUGCACAUGUGUGUGUGCGUGUAGAGAGAAAU ......((((((-----((((....................((((((((..............................))))))))(((((((((....))))))))).)))))))))) ( -26.41) >DroSim_CAF1 40764 115 + 1 GUUUUCAUUUCU-----UUUUUUUUAGCCCCUUCCCCCAAUGGCAUGCUGCAUGUCUUCAUUAUCGUUUAUUGAAUACAAGCAUGCUUAUGCACAUGUGUGUGUGCGUGUAGAGAGAAAU ......((((((-----((((....................((((((((...(((.((((.((.....)).)))).)))))))))))(((((((((....))))))))).)))))))))) ( -32.10) >DroEre_CAF1 40937 103 + 1 GUUUUCAUUUCU-----UU-UUUUUAGCCCCUUCCCCCAAUGGCAUGCUGCAUGUCUUCAUUAUCGUUC----AAUACAAGCAUGCUUAUAC----AUAUG--UGUCUGUAGAGAGA-AU ........((((-----((-((..(((..............((((((((..(((....)))....((..----...)).)))))))).((((----....)--)))))).)))))))-). ( -16.80) >DroYak_CAF1 39971 110 + 1 GUUUUCAUUUCU-----UU--UUUUAGCCCCUUCCCCCAAUGGCAUGCUGCAUGUCUUCAUUAUCGUUUAUUGAAUACAAGCAUGCUUAUACAUACAUAUG--UGUCUGUAGAGAGA-AU ........((((-----((--((.(((..............((((((((...(((.((((.((.....)).)))).))))))))))).((((((....)))--)))))).)))))))-). ( -24.80) >consensus GUUUUCAUUUCU_____UUUUUUUUAGCCCCUUCCCCCAAUGGCAUGCUGCAUGUCUUCAUUAUCGUUUAUUGAAUACAAGCAUGCUUAUACACAUGUAUGUGUGCGUGUAGAGAGA_AU .(((((...................................((((((((...(((.((((.((.....)).)))).)))))))))))(((((((((....)))))))))..))))).... (-18.98 = -21.04 + 2.06)


| Location | 4,437,500 – 4,437,619 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.43 |
| Mean single sequence MFE | -24.63 |
| Consensus MFE | -20.44 |
| Energy contribution | -20.98 |
| Covariance contribution | 0.54 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.955501 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4437500 119 - 22224390 AU-UCUCUCUACACGCACACAUACAUGUGUAUAAGCAUGCUUGUAUUCAAUAAACGAUAAUGAAGACAUGCAGCAUGCCAUUGGGGGAAGGGGCUAAAAAAAAAAAGAAGAAAUGAAAAC .(-((((((.....(((((......)))))....((((((((((.((((.((.....)).)))).)))...)))))))....)))))))............................... ( -25.10) >DroSec_CAF1 39332 115 - 1 AUUUCUCUCUACACGCACACACACAUGUGCAUAAGCAUGCUUGUAUUUAAUAAACGAUAAUGAAGACAUGCAGCAUGCCAUUGGGGGAAGGGGCUAAAAAAAA-----AGAAAUGAAAAC .((((((((.....(((((......)))))....((((((((((.((((.((.....)).)))).)))...)))))))....)))))))).............-----............ ( -25.90) >DroSim_CAF1 40764 115 - 1 AUUUCUCUCUACACGCACACACACAUGUGCAUAAGCAUGCUUGUAUUCAAUAAACGAUAAUGAAGACAUGCAGCAUGCCAUUGGGGGAAGGGGCUAAAAAAAA-----AGAAAUGAAAAC .((((((((.....(((((......)))))....((((((((((.((((.((.....)).)))).)))...)))))))....)))))))).............-----............ ( -28.10) >DroEre_CAF1 40937 103 - 1 AU-UCUCUCUACAGACA--CAUAU----GUAUAAGCAUGCUUGUAUU----GAACGAUAAUGAAGACAUGCAGCAUGCCAUUGGGGGAAGGGGCUAAAAA-AA-----AGAAAUGAAAAC .(-((((((((((....--....)----)))...(((((((.((((.----................)))))))))))....)))))))...........-..-----............ ( -20.13) >DroYak_CAF1 39971 110 - 1 AU-UCUCUCUACAGACA--CAUAUGUAUGUAUAAGCAUGCUUGUAUUCAAUAAACGAUAAUGAAGACAUGCAGCAUGCCAUUGGGGGAAGGGGCUAAAA--AA-----AGAAAUGAAAAC .(-((((((((((.(((--....))).))))...((((((((((.((((.((.....)).)))).)))...)))))))....)))))))..........--..-----............ ( -23.90) >consensus AU_UCUCUCUACACGCACACAUACAUGUGUAUAAGCAUGCUUGUAUUCAAUAAACGAUAAUGAAGACAUGCAGCAUGCCAUUGGGGGAAGGGGCUAAAAAAAA_____AGAAAUGAAAAC ...((((((.....(((((......)))))....((((((((((.((((.((.....)).)))).)))...)))))))....))))))................................ (-20.44 = -20.98 + 0.54)

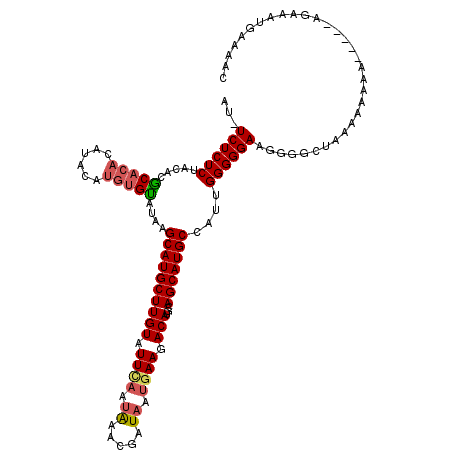
| Location | 4,437,540 – 4,437,659 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.67 |
| Mean single sequence MFE | -20.09 |
| Consensus MFE | -16.06 |
| Energy contribution | -16.60 |
| Covariance contribution | 0.54 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908081 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4437540 119 - 22224390 CGGUAGAAUACCCCCCCCCCCCCCACCACCACUUUUAUAUAU-UCUCUCUACACGCACACAUACAUGUGUAUAAGCAUGCUUGUAUUCAAUAAACGAUAAUGAAGACAUGCAGCAUGCCA (((((((...................................-....))))).)).(((((....)))))....((((((((((.((((.((.....)).)))).)))...))))))).. ( -19.93) >DroSec_CAF1 39367 105 - 1 CGGUAGAAUACCCCCCAC---------------UUUAUAUAUUUCUCUCUACACGCACACACACAUGUGCAUAAGCAUGCUUGUAUUUAAUAAACGAUAAUGAAGACAUGCAGCAUGCCA ..(((((...........---------------..............)))))..(((((......)))))....((((((((((.((((.((.....)).)))).)))...))))))).. ( -20.31) >DroSim_CAF1 40799 105 - 1 CGGUAGAAUACCCCCCAC---------------UUUAUAUAUUUCUCUCUACACGCACACACACAUGUGCAUAAGCAUGCUUGUAUUCAAUAAACGAUAAUGAAGACAUGCAGCAUGCCA ..(((((...........---------------..............)))))..(((((......)))))....((((((((((.((((.((.....)).)))).)))...))))))).. ( -22.51) >DroEre_CAF1 40971 95 - 1 CGGUAGAAUACCCCCCUC--------------UUUUAUAUAU-UCUCUCUACAGACA--CAUAU----GUAUAAGCAUGCUUGUAUU----GAACGAUAAUGAAGACAUGCAGCAUGCCA .((((.............--------------..((((((((-....((....))..--...))----))))))((((((((.((((----....))))...))).)))))....)))). ( -17.80) >DroYak_CAF1 40004 96 - 1 CGGUAGAAUACCCCCU---------------------CAUAU-UCUCUCUACAGACA--CAUAUGUAUGUAUAAGCAUGCUUGUAUUCAAUAAACGAUAAUGAAGACAUGCAGCAUGCCA .(..((((((......---------------------..)))-)))..)((((.(((--....))).))))...((((((((((.((((.((.....)).)))).)))...))))))).. ( -19.90) >consensus CGGUAGAAUACCCCCCAC_______________UUUAUAUAU_UCUCUCUACACGCACACAUACAUGUGUAUAAGCAUGCUUGUAUUCAAUAAACGAUAAUGAAGACAUGCAGCAUGCCA ..(((((........................................)))))..(((((......)))))....((((((((((.((((.((.....)).)))).)))...))))))).. (-16.06 = -16.60 + 0.54)



| Location | 4,437,580 – 4,437,699 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.03 |
| Mean single sequence MFE | -23.24 |
| Consensus MFE | -19.58 |
| Energy contribution | -19.80 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938111 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4437580 119 - 22224390 UCGGCAACGCGUGCAAUACUCAGGGAUUACCUUUGUCUCCCGGUAGAAUACCCCCCCCCCCCCCACCACCACUUUUAUAUAU-UCUCUCUACACGCACACAUACAUGUGUAUAAGCAUGC ..(....)((((((..((((..(((((.......)))))..)))).....................................-...........(((((......)))))....)))))) ( -21.30) >DroSec_CAF1 39407 105 - 1 UCGGCAACGCGUGCAAUACUCAGGGAUUACCUUUGUCUCCCGGUAGAAUACCCCCCAC---------------UUUAUAUAUUUCUCUCUACACGCACACACACAUGUGCAUAAGCAUGC ..(....)((((((........((((...........)))).(((((...........---------------..............)))))..(((((......)))))....)))))) ( -24.01) >DroSim_CAF1 40839 105 - 1 UCGGCAACGCGUGCAAUACUCAGGGAUUACCUUUGUCUCCCGGUAGAAUACCCCCCAC---------------UUUAUAUAUUUCUCUCUACACGCACACACACAUGUGCAUAAGCAUGC ..(....)((((((........((((...........)))).(((((...........---------------..............)))))..(((((......)))))....)))))) ( -24.01) >DroEre_CAF1 41007 99 - 1 UCGGCAACGCGUGCAAUACUGAGGGAUUACCUUUGUCUCCCGGUAGAAUACCCCCCUC--------------UUUUAUAUAU-UCUCUCUACAGACA--CAUAU----GUAUAAGCAUGC ..(....)((((((..(((((.(((((.......))))).))))).............--------------..((((((((-....((....))..--...))----)))))))))))) ( -23.70) >DroYak_CAF1 40044 96 - 1 UCGGCAACGCGUGCAAUACUCAGGGAUUACCUUUGCCUCCCGGUAGAAUACCCCCU---------------------CAUAU-UCUCUCUACAGACA--CAUAUGUAUGUAUAAGCAUGC ..(....)((((((.......((((..((..((((((....)))))).))..))))---------------------.....-......((((.(((--....))).))))...)))))) ( -23.20) >consensus UCGGCAACGCGUGCAAUACUCAGGGAUUACCUUUGUCUCCCGGUAGAAUACCCCCCAC_______________UUUAUAUAU_UCUCUCUACACGCACACAUACAUGUGUAUAAGCAUGC ..(....)((((((........((((...........)))).(((((........................................)))))..(((((......)))))....)))))) (-19.58 = -19.80 + 0.22)

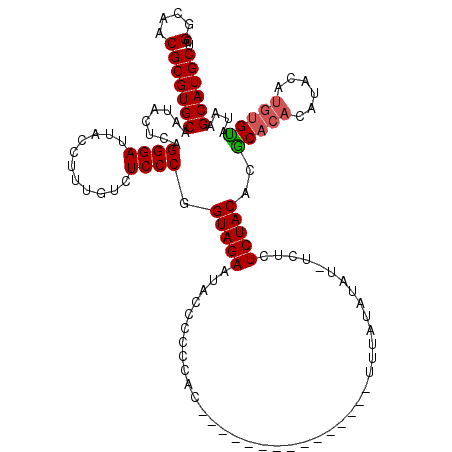
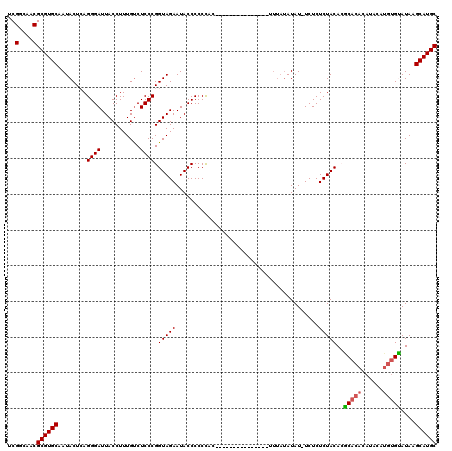
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:12:05 2006