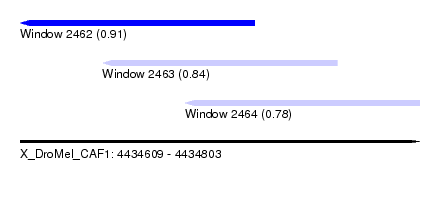
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,434,609 – 4,434,803 |
| Length | 194 |
| Max. P | 0.909501 |

| Location | 4,434,609 – 4,434,723 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.64 |
| Mean single sequence MFE | -27.80 |
| Consensus MFE | -23.50 |
| Energy contribution | -23.75 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.909501 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4434609 114 - 22224390 CAUAUAUAUAUGUACUAUAUGUGGUCGUCUAUA------UAUAUGUACACGUGUUUAUGGCAAUACCUGAACCUGUGCUCUCUUUUUUCUCUGGCCAUCAACGCACGCGCCCGCAUUUAG ..........((((((((((((((....)))))------)))).))))).(((((.(((((.......(((...(......)....)))....))))).)))))..((....))...... ( -26.00) >DroSec_CAF1 37014 118 - 1 CAUAUAUAUAUGUACUAUAUGUGGUCUGCUAUAUAUA--UAUAUAUACACGUGUUUAUGGCAAUACCUGAACCUGUGCUCUCUUUUUUCUCUGGCCAUCAACGCACGCGCCCGCAUUUAG ......((((((((.(((((((((....)))))))))--))))))))...(((((.(((((.......(((...(......)....)))....))))).)))))..((....))...... ( -26.40) >DroEre_CAF1 37978 118 - 1 CAUAUACAUA--UACUGUAUGUGGUCGUCUAUAUAUAUAUAUAUGUACACGUGUUUAUGGCAAUACCUGAACCUGUGCUCUCUUUUUUCUCUGGCCAUCAACGCACGCGCCCGCAUUUAG ....((((((--((.(((((((((....)))))))))..))))))))...(((((.(((((.......(((...(......)....)))....))))).)))))..((....))...... ( -26.30) >DroYak_CAF1 37083 114 - 1 CAUAU----A--UACUAUAUGCGGUCGUCUGUAUAUACAUGUGUGUACACGUGUUUAUGGCAAUACCUGAACCUGUGCUCUCUUUUUUCUCUGGCCAUCAACGCACGCGCCCGCAUUUAG .....----.--..(((.((((((.(((.((((((((....)))))))).(((((.(((((.......(((...(......)....)))....))))).)))))..))).)))))).))) ( -32.50) >consensus CAUAUAUAUA__UACUAUAUGUGGUCGUCUAUAUAUA__UAUAUGUACACGUGUUUAUGGCAAUACCUGAACCUGUGCUCUCUUUUUUCUCUGGCCAUCAACGCACGCGCCCGCAUUUAG ..............(((.((((((.(((.(((((((....)))))))...(((((.(((((.......(((...(......)....)))....))))).)))))..))).)))))).))) (-23.50 = -23.75 + 0.25)

| Location | 4,434,649 – 4,434,763 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.85 |
| Mean single sequence MFE | -29.92 |
| Consensus MFE | -20.39 |
| Energy contribution | -21.83 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.844089 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4434649 114 - 22224390 CUCGGCUUCCACUGUGUCCAUCUGGAUGUAUAAAAUGCACCAUAUAUAUAUGUACUAUAUGUGGUCGUCUAUA------UAUAUGUACACGUGUUUAUGGCAAUACCUGAACCUGUGCUC ...((.(((...((..(((....)))..)).........(((((.(((..((((((((((((((....)))))------)))).)))))..))).)))))........)))))....... ( -29.70) >DroSec_CAF1 37054 118 - 1 CUCCUUUUCCACUGUGUCCAUCUGGAUGUAUAAAAUGCACCAUAUAUAUAUGUACUAUAUGUGGUCUGCUAUAUAUA--UAUAUAUACACGUGUUUAUGGCAAUACCUGAACCUGUGCUC ............((..(((....)))..))......((((......((((((((.(((((((((....)))))))))--))))))))...(.(((((.((.....)))))))).)))).. ( -26.80) >DroEre_CAF1 38018 118 - 1 CUCCGUGUCCACUGUGUCCAUCUGGAUGUAUAAAGUGCACCAUAUACAUA--UACUGUAUGUGGUCGUCUAUAUAUAUAUAUAUGUACACGUGUUUAUGGCAAUACCUGAACCUGUGCUC ..(((((..(((.((((.(((.((.(((((((.((((.((((((((((..--...)))))))))))).)).))))))).)).))).)))))))..))))).................... ( -34.60) >DroYak_CAF1 37123 114 - 1 CUCCAUUUCCACAGUGUCCAUCUGGAUGUAUAAAAUGCACCAUAU----A--UACUAUAUGCGGUCGUCUGUAUAUACAUGUGUGUACACGUGUUUAUGGCAAUACCUGAACCUGUGCUC .........(((((.((...((.((.(((((...)))))(((((.----(--((((((((((((....)))))))))...(((....))))))).))))).....)).)))))))))... ( -28.60) >consensus CUCCGUUUCCACUGUGUCCAUCUGGAUGUAUAAAAUGCACCAUAUAUAUA__UACUAUAUGUGGUCGUCUAUAUAUA__UAUAUGUACACGUGUUUAUGGCAAUACCUGAACCUGUGCUC .........(((.((((.(((.((.(((((((...((.((((((((((.......))))))))))))..)))))))...)).))).))))(.(((((.((.....)))))))).)))... (-20.39 = -21.83 + 1.44)

| Location | 4,434,689 – 4,434,803 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.66 |
| Mean single sequence MFE | -28.20 |
| Consensus MFE | -18.62 |
| Energy contribution | -21.37 |
| Covariance contribution | 2.75 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.776832 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4434689 114 - 22224390 AUGAGCCCUCUUCCAGGCCAGGUCCAUUGGAAGCCUUUUUCUCGGCUUCCACUGUGUCCAUCUGGAUGUAUAAAAUGCACCAUAUAUAUAUGUACUAUAUGUGGUCGUCUAUA------U ...........((((((...((.(((.((((((((........)))))))).)).).)).)))))).(((((..(((.((((((((((.......))))))))))))).))))------) ( -37.10) >DroSec_CAF1 37094 118 - 1 AUGAGCCCUCUUCCAGGGCAGGUCCAUUGGUAGCCUUUUCCUCCUUUUCCACUGUGUCCAUCUGGAUGUAUAAAAUGCACCAUAUAUAUAUGUACUAUAUGUGGUCUGCUAUAUAUA--U ....(((((.....)))))..........(((((..................((..(((....)))..))........((((((((((.......))))))))))..))))).....--. ( -27.80) >DroEre_CAF1 38058 118 - 1 AUGAACCAUUUUCCAGGCCAGGUCCAUUGAAAGCCUUUGCCUCCGUGUCCACUGUGUCCAUCUGGAUGUAUAAAGUGCACCAUAUACAUA--UACUGUAUGUGGUCGUCUAUAUAUAUAU ...........(((((((((((..(((.((..((....)).)).)))..).))).))....)))))((((((.((((.((((((((((..--...)))))))))))).)).))))))... ( -30.10) >DroYak_CAF1 37163 114 - 1 AUGAACCACCUUUUAGGCCAGGUCCAUUAAAAGCCUUUUCCUCCAUUUCCACAGUGUCCAUCUGGAUGUAUAAAAUGCACCAUAU----A--UACUAUAUGCGGUCGUCUGUAUAUACAU (((.(((.((.....))...))).))).........................(((((..((.(((.(((((...)))))))).))----)--))))((((((((....)))))))).... ( -17.80) >consensus AUGAACCAUCUUCCAGGCCAGGUCCAUUGAAAGCCUUUUCCUCCGUUUCCACUGUGUCCAUCUGGAUGUAUAAAAUGCACCAUAUAUAUA__UACUAUAUGUGGUCGUCUAUAUAUA__U ...........((((((...((.(((.(((((((..........))))))).)).).)).))))))((((((...((.((((((((((.......))))))))))))..))))))..... (-18.62 = -21.37 + 2.75)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:12:00 2006