
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,434,252 – 4,434,530 |
| Length | 278 |
| Max. P | 0.999990 |

| Location | 4,434,252 – 4,434,370 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.01 |
| Mean single sequence MFE | -37.00 |
| Consensus MFE | -31.07 |
| Energy contribution | -31.87 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.963768 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4434252 118 + 22224390 UGGGAU--AUGGGUUUUGUGGUAUUAGAUCUAUGAUCUGGGAUGGGUAAUAUCAUUUGGUGGGUGUGAGGGGCUGGGGAGGUAGAUACCUGCGGGCGCGGAUGUUGCGACCCUUUUCCCA .((((.--..(((((........(((((((...))))))).....((((((((..((.(..(((((.....(((.....)))..)))))..).))....)))))))))))))...)))). ( -38.20) >DroSec_CAF1 36656 120 + 1 UGGGAUAUAUGGGUUUUGUGGUAUUAGAUCUAUGAUCUGGGAUGGGUAAUAACAUUUGGUGGGUGUGAGGGGCAGGGGAGGAAGAUACCUGCGGGCGCGGAUGUUGCGACCCUUUUCCCA ..((((.(((((((((.........)))))))))))))((((.((((..(((((((.......((((....(((((...........)))))...)))))))))))..))))...)))). ( -37.51) >DroSim_CAF1 38025 120 + 1 UGGGAUAUAUGGGUUUUGUGGUAUUAGAUCUAUGAUCUGGGAUGGGUAAUAACAUUUGGUGGGUGUGAGGGGCAGGGGAGGAAGAUACCUGCGGGCGCGGAUGUUGCGACCCUUUUCCCA ..((((.(((((((((.........)))))))))))))((((.((((..(((((((.......((((....(((((...........)))))...)))))))))))..))))...)))). ( -37.51) >DroEre_CAF1 37640 113 + 1 UGGGAU--AUGGGUUUUGUGGUAUUAGAUCUAUGAUCUGGGAUGGGUAAUAUCAUAUAGUGGGUGUGAGGAACAG-----GGAGAUACCUGCGGGCGCGGAUGUUGCGACCCUUUUCCCA .((((.--..(((((........(((((((...))))))).....((((((((.....(..(((((.........-----....)))))..).......)))))))))))))...)))). ( -35.92) >DroYak_CAF1 36740 113 + 1 UGGGAU--AUGGGUUUUGUGGUAUUAGAUCUAUGAUCUGGGAUGGGUAAUAUCAUUUUGUGGGUGGGAGAAAAAG-----GGAGAUACCUGCGGGCGCGGAUGUUGCGACCCUUUUCCCA .((((.--..(((((........(((((((...))))))).....((((((((..((((..((((..........-----.....))))..))))....)))))))))))))...)))). ( -35.86) >consensus UGGGAU__AUGGGUUUUGUGGUAUUAGAUCUAUGAUCUGGGAUGGGUAAUAUCAUUUGGUGGGUGUGAGGGGCAGGGGAGGAAGAUACCUGCGGGCGCGGAUGUUGCGACCCUUUUCCCA .((((.....(((((........(((((((...))))))).....((((((((..((.(..(((((..................)))))..).))....)))))))))))))...)))). (-31.07 = -31.87 + 0.80)

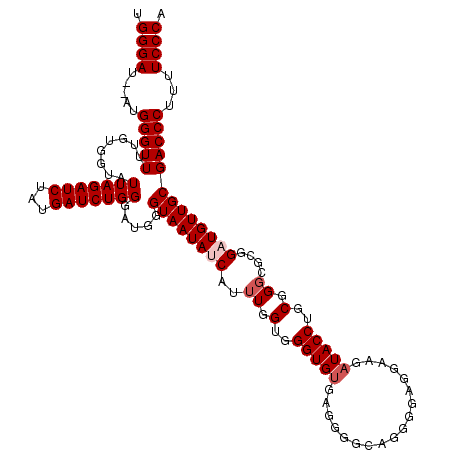

| Location | 4,434,252 – 4,434,370 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.01 |
| Mean single sequence MFE | -24.30 |
| Consensus MFE | -22.88 |
| Energy contribution | -22.88 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.48 |
| Structure conservation index | 0.94 |
| SVM decision value | 5.56 |
| SVM RNA-class probability | 0.999990 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4434252 118 - 22224390 UGGGAAAAGGGUCGCAACAUCCGCGCCCGCAGGUAUCUACCUCCCCAGCCCCUCACACCCACCAAAUGAUAUUACCCAUCCCAGAUCAUAGAUCUAAUACCACAAAACCCAU--AUCCCA .((((...(((.(((.......))))))...(((((.............................(((........)))...(((((...))))).)))))...........--.)))). ( -23.10) >DroSec_CAF1 36656 120 - 1 UGGGAAAAGGGUCGCAACAUCCGCGCCCGCAGGUAUCUUCCUCCCCUGCCCCUCACACCCACCAAAUGUUAUUACCCAUCCCAGAUCAUAGAUCUAAUACCACAAAACCCAUAUAUCCCA .((((...(((.(((.......))))))(((((...........))))).............................))))(((((...)))))......................... ( -25.40) >DroSim_CAF1 38025 120 - 1 UGGGAAAAGGGUCGCAACAUCCGCGCCCGCAGGUAUCUUCCUCCCCUGCCCCUCACACCCACCAAAUGUUAUUACCCAUCCCAGAUCAUAGAUCUAAUACCACAAAACCCAUAUAUCCCA .((((...(((.(((.......))))))(((((...........))))).............................))))(((((...)))))......................... ( -25.40) >DroEre_CAF1 37640 113 - 1 UGGGAAAAGGGUCGCAACAUCCGCGCCCGCAGGUAUCUCC-----CUGUUCCUCACACCCACUAUAUGAUAUUACCCAUCCCAGAUCAUAGAUCUAAUACCACAAAACCCAU--AUCCCA .((((...(((.(((.......))))))(((((......)-----)))).............................))))(((((...))))).................--...... ( -24.50) >DroYak_CAF1 36740 113 - 1 UGGGAAAAGGGUCGCAACAUCCGCGCCCGCAGGUAUCUCC-----CUUUUUCUCCCACCCACAAAAUGAUAUUACCCAUCCCAGAUCAUAGAUCUAAUACCACAAAACCCAU--AUCCCA .((((...(((.(((.......))))))...(((((....-----....................(((........)))...(((((...))))).)))))...........--.)))). ( -23.10) >consensus UGGGAAAAGGGUCGCAACAUCCGCGCCCGCAGGUAUCUCCCUCCCCUGCCCCUCACACCCACCAAAUGAUAUUACCCAUCCCAGAUCAUAGAUCUAAUACCACAAAACCCAU__AUCCCA .((((...(((.(((.......))))))...(((((.............................(((........)))...(((((...))))).)))))..............)))). (-22.88 = -22.88 + 0.00)

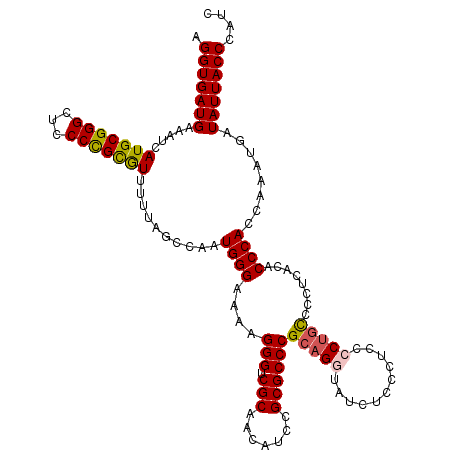
| Location | 4,434,290 – 4,434,410 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.05 |
| Mean single sequence MFE | -36.24 |
| Consensus MFE | -32.88 |
| Energy contribution | -32.96 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.91 |
| SVM decision value | 3.73 |
| SVM RNA-class probability | 0.999570 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4434290 120 - 22224390 AGGUGAUGAAAUCAUGCGGGCUCCCCGCGUUUUUAGCCAAUGGGAAAAGGGUCGCAACAUCCGCGCCCGCAGGUAUCUACCUCCCCAGCCCCUCACACCCACCAAAUGAUAUUACCCAUC .((((.(((....(((((((...))))))).....((...((((....(((.(((.......))))))..((((....)))).))))))...)))))))..................... ( -34.80) >DroSec_CAF1 36696 120 - 1 AGGUGAUGAAAUCAUGCGGGCUCCCCGCGUUUUUAGCCAAUGGGAAAAGGGUCGCAACAUCCGCGCCCGCAGGUAUCUUCCUCCCCUGCCCCUCACACCCACCAAAUGUUAUUACCCAUC .((((((((....(((((((...)))))))..........((((....(((.(((.......))))))(((((...........)))))........)))).......)))))))).... ( -39.40) >DroSim_CAF1 38065 120 - 1 AGGUGAUGAAAUCAUGCGGGCUCCCCGCGUUUUUAGCCAAUGGGAAAAGGGUCGCAACAUCCGCGCCCGCAGGUAUCUUCCUCCCCUGCCCCUCACACCCACCAAAUGUUAUUACCCAUC .((((((((....(((((((...)))))))..........((((....(((.(((.......))))))(((((...........)))))........)))).......)))))))).... ( -39.40) >DroEre_CAF1 37678 115 - 1 AGGUGAUGAAAUCAUGCGGGCUCCCCGUGUUUUUAGCCAAUGGGAAAAGGGUCGCAACAUCCGCGCCCGCAGGUAUCUCC-----CUGUUCCUCACACCCACUAUAUGAUAUUACCCAUC .((((((...((((((((((...)))))............((((....(((.(((.......))))))(((((......)-----))))........))))....))))))))))).... ( -35.40) >DroYak_CAF1 36778 115 - 1 AGGUGAUGAAAUCAUGCGGGCUCCCUGCAUUUUUAGCCAAUGGGAAAAGGGUCGCAACAUCCGCGCCCGCAGGUAUCUCC-----CUUUUUCUCCCACCCACAAAAUGAUAUUACCCAUC .((((((...(((((((((((((((.((.......))....))))...((((......))))..))))))(((......)-----))..................))))))))))).... ( -32.20) >consensus AGGUGAUGAAAUCAUGCGGGCUCCCCGCGUUUUUAGCCAAUGGGAAAAGGGUCGCAACAUCCGCGCCCGCAGGUAUCUCCCUCCCCUGCCCCUCACACCCACCAAAUGAUAUUACCCAUC .(((((((.....(((((((...)))))))..........((((....(((.(((.......))))))(((((...........)))))........))))........))))))).... (-32.88 = -32.96 + 0.08)

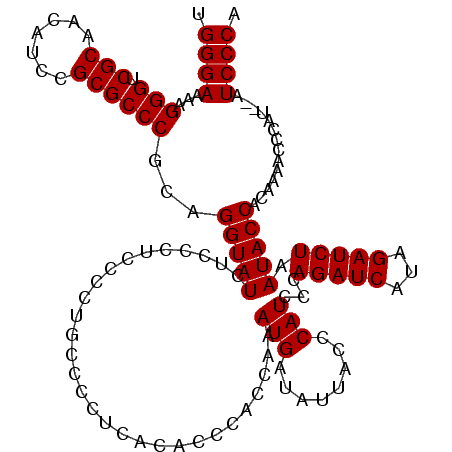
| Location | 4,434,410 – 4,434,530 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.64 |
| Mean single sequence MFE | -36.14 |
| Consensus MFE | -32.14 |
| Energy contribution | -32.34 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.89 |
| SVM decision value | 4.06 |
| SVM RNA-class probability | 0.999780 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4434410 120 + 22224390 UAUGUAAUCAUCAAAUGAAGAGGGGACCUCGAAUUGGAUGCAAAGUGGAGGCCUCCACGGCAGACAAAUCAAUAAAAUGUCUGUAUCUGUAUCUGGUUGGUUCUCCCAGUUCUUUUUUUU ............(((.(((((.((((...(.(((..((((((..(((((....))))).(((((((...........)))))))...))))))..))).)...))))...))))).))). ( -38.30) >DroSec_CAF1 36816 119 + 1 UAUGUAAUCAUCAAAUGAAGAGGGAACCUCGAAUUGGAUGCAAAGUGGAGGCCCUCACGGCAGACAAAUCAAUAAAAUGUCUGUAUCUGUAUCUGGUUGGUUCUCCCAGUUCUUU-UUCU ................((((((((((((...(((..((((((..((((......)))).(((((((...........)))))))...))))))..)))))))))).....)))))-.... ( -31.90) >DroSim_CAF1 38185 120 + 1 UAUGUAAUCAUCAAAUGAAGAGGGGACUGCGAAUUGGAUUGAAAGUGGAGGCCUCCACGGCAGACAAAUCAAUAAAAUGUCUGUAUCAGUAUCUGGUUGGUUCUCCCAGUUCUUUUUUCU ................(((((((((((((..(((..((((((..(((((....))))).(((((((...........))))))).)))..)))..)))((....))))))))))))))). ( -36.70) >DroEre_CAF1 37793 115 + 1 UAUGUAAUCAUCAAAUGAAGAGGGAACCUCGAAUUGGAUGCAAAGUGGAGGCCUUCACGGCAGACAAAUCAAUAAAAUGUCUGUAUCUGUAUCUGGUUGGUUCUCCUUGUGUUUU----- ...................(((((((((...(((..((((((..(((((....))))).(((((((...........)))))))...))))))..)))))))).)))).......----- ( -34.80) >DroYak_CAF1 36893 117 + 1 UAUGUAAUCAUCAACUGAAGAGGGGACCUCGAAUUGGAUGCAAAGUGGAGGCCUCCACGACAGACAAAUCAAUAAAAUGUCUGUAUCUGUAUCUGGUCGGUUCUCUCAGUUUUUUUU--- ...................(((((((((....((..((((((..(((((....))))).(((((((...........)))))))...))))))..)).)))))))))..........--- ( -39.00) >consensus UAUGUAAUCAUCAAAUGAAGAGGGGACCUCGAAUUGGAUGCAAAGUGGAGGCCUCCACGGCAGACAAAUCAAUAAAAUGUCUGUAUCUGUAUCUGGUUGGUUCUCCCAGUUCUUUUUU_U .......((.((....)).))(((((((...(((..((((((..(((((....))))).(((((((...........)))))))...))))))..))))))))))............... (-32.14 = -32.34 + 0.20)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:11:57 2006