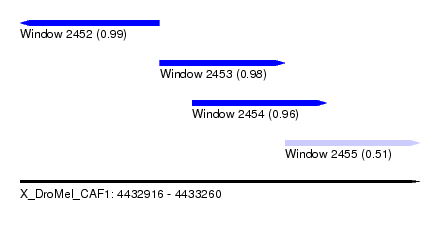
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,432,916 – 4,433,260 |
| Length | 344 |
| Max. P | 0.994656 |

| Location | 4,432,916 – 4,433,036 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.00 |
| Mean single sequence MFE | -37.28 |
| Consensus MFE | -37.76 |
| Energy contribution | -37.28 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.77 |
| Structure conservation index | 1.01 |
| SVM decision value | 2.50 |
| SVM RNA-class probability | 0.994656 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4432916 120 - 22224390 AGGCCAGAGACCAUAUCACGCGCCUUCGUUUGGCGGGGGGCUUAAAAAUCGCUUCCAGCGUCUUUGAUACUUUGGAACCAAAGCCCAGAGACAACAGCAAGAGGCUCCACAGAUAUAUCU .......(((..(((((...((((.......)))).(((((((.......((.....))(((((((...(((((....)))))..))))))).........)))))))...))))).))) ( -37.00) >DroSec_CAF1 35273 120 - 1 AGGCCAGAGACCAUAUCACGCGCCUUCGUUUGGCGGGGGGCUUAAAAAUCGCUUCCAGCGUCUUUGAUACUUUGGAACCAAAGCCCAGAGACAACAGCAAGAGGCUCCACAGAUAUAUCU .......(((..(((((...((((.......)))).(((((((.......((.....))(((((((...(((((....)))))..))))))).........)))))))...))))).))) ( -37.00) >DroSim_CAF1 36631 120 - 1 AGGCCAGAGACCAUAUCACGCGCCUUCGUUUGGCGGGGGGCUUAAAAAUCGCUUCCAGCGUCUUUGAUACUUUGGAACCAAAGCCCAGAGACAACAGCAAGAGGCUCCACAGAUAUAUCU .......(((..(((((...((((.......)))).(((((((.......((.....))(((((((...(((((....)))))..))))))).........)))))))...))))).))) ( -37.00) >DroEre_CAF1 36211 120 - 1 AGGCCAGAGACCGUAUCACGCGCCUUCGUUUGGCGAGGGGCUUAAAAAUCGCUUCCAGCGUCUUUGAUACUUUGGAACCAAAGCCCAGAGACAACAGCAAGAAGCUCCACAGAUAUAUCU .......(((..(((((...((((.......)))).(((((((.......((.....))(((((((...(((((....)))))..))))))).........)))))))...))))).))) ( -37.60) >DroYak_CAF1 35410 120 - 1 AGGCCAGAGACCAUAUCACGCGCCUUCGUUUGGCGGGGAGCUUAAAAAUCGCUUCCAGCGUCUUUGAUACUUUGGAAGCAAAGCCCAGAGACAACAGCAAAAGGCUCCACAGAUAUAUCU .......(((..(((((...((((.......)))).(((((((.......((.....))(((((((...(((((....)))))..))))))).........)))))))...))))).))) ( -37.80) >consensus AGGCCAGAGACCAUAUCACGCGCCUUCGUUUGGCGGGGGGCUUAAAAAUCGCUUCCAGCGUCUUUGAUACUUUGGAACCAAAGCCCAGAGACAACAGCAAGAGGCUCCACAGAUAUAUCU .......(((..(((((...((((.......)))).(((((((.......((.....))(((((((...(((((....)))))..))))))).........)))))))...))))).))) (-37.76 = -37.28 + -0.48)

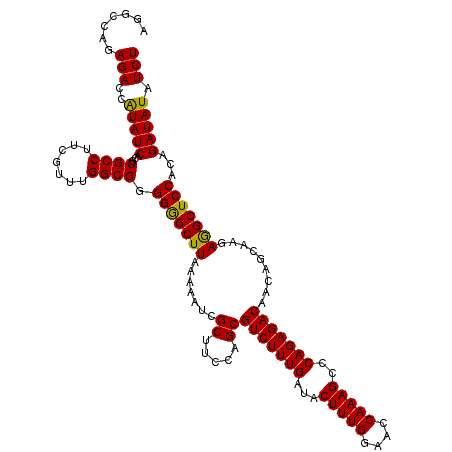
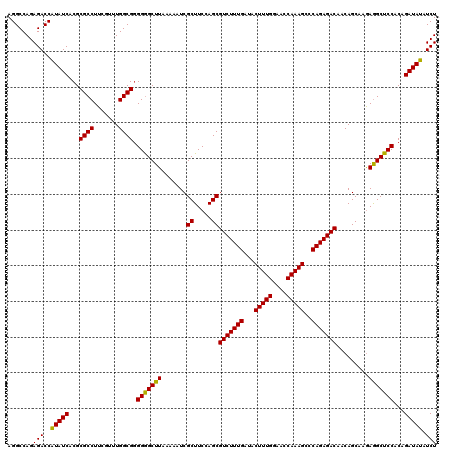
| Location | 4,433,036 – 4,433,144 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.06 |
| Mean single sequence MFE | -18.78 |
| Consensus MFE | -17.16 |
| Energy contribution | -17.56 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.88 |
| SVM RNA-class probability | 0.981215 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4433036 108 + 22224390 UCCUCCGAUCAAUUCCCAUUCCC------------AUUGCCAUUCCCAUACCGAUUUCGAUAGAUUUCGCCUUGUGUGAUCGCUAUCACGCACUCCUCCUCCGGCUGAUUCAGCUGAUUC .......................------------................(((..((....))..)))...((((((((....)))))))).........((((((...)))))).... ( -19.10) >DroSec_CAF1 35393 120 + 1 UCCUCCGAUCAAUUCCCAUUCCCAUUCCCAUUCUCAUUGCCAUUCCCAUACCGAUUUCGAUAGAUUUCGCCUUGUGUGAUCGCUAUCACGCACUCCUCCUCCGGCUGAUUCAGCUGAUUC ...................................................(((..((....))..)))...((((((((....)))))))).........((((((...)))))).... ( -19.10) >DroSim_CAF1 36751 114 + 1 UCCUCCGAUCAAUUCCCAUUCCCAUUCCC------AUUGCCAUUCCCAUACCGAUUUCGAUAGAUUUCGCCUUGUGUGAUCGCUAUCACGCACUCCUCCUCCGGCUGAUUCAGCUGAUUC .............................------................(((..((....))..)))...((((((((....)))))))).........((((((...)))))).... ( -19.10) >DroEre_CAF1 36331 114 + 1 UCCUCCGAUCAAUUCCCAUUCCCAUUUCC------AUUGCCAUUCCCAUUCCGAUUUCGAUAGAUUUCGCCUUAUGUGAUCGCAAUCACGCACUCCUCCUCCGGCUGAUUCAGCUGAUUC .............................------(((((...((.(((..(((..((....))..)))....))).))..)))))...............((((((...)))))).... ( -17.20) >DroYak_CAF1 35530 108 + 1 UCCUCCGAUCGAUUCCCAUUCCC------------AUUGCCAUUUCCAUUCCGAUUUCGAUAGAUUUCGCCUUGUGUGAUCGCUAUCACGCACUCCUCCUCCGGCUGGUUCAGCUUAUUC .....................((------------(..(((..........(((..((....))..)))...((((((((....))))))))..........))))))............ ( -19.40) >consensus UCCUCCGAUCAAUUCCCAUUCCCAUU_CC______AUUGCCAUUCCCAUACCGAUUUCGAUAGAUUUCGCCUUGUGUGAUCGCUAUCACGCACUCCUCCUCCGGCUGAUUCAGCUGAUUC ...................................................(((..((....))..)))...((((((((....)))))))).........((((((...)))))).... (-17.16 = -17.56 + 0.40)

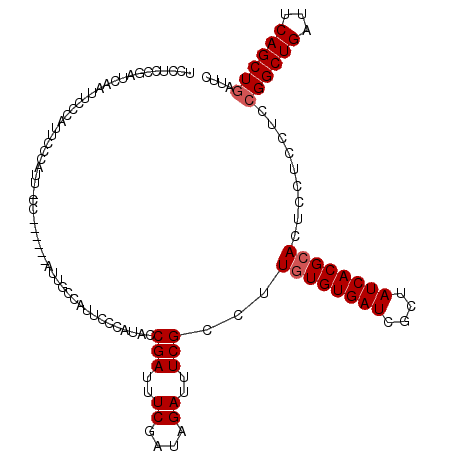
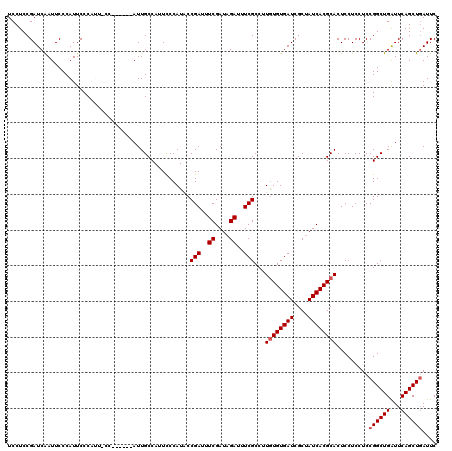
| Location | 4,433,064 – 4,433,180 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.77 |
| Mean single sequence MFE | -29.92 |
| Consensus MFE | -24.36 |
| Energy contribution | -24.56 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.963413 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4433064 116 + 22224390 CAUUCCCAUACCGAUUUCGAUAGAUUUCGCCUUGUGUGAUCGCUAUCACGCACUCCUCCUCCGGCUGAUUCAGCUGAUUCCGGCCAGGUGACUCACCC----GCAAUCAUCACUCUUGGC ..........((((....(((.((((.((((.((((((((....)))))))).........((((((...)))))).....)))..((((...)))).----).)))))))....)))). ( -29.20) >DroSec_CAF1 35433 120 + 1 CAUUCCCAUACCGAUUUCGAUAGAUUUCGCCUUGUGUGAUCGCUAUCACGCACUCCUCCUCCGGCUGAUUCAGCUGAUUCCGGCCAGGUGACUCACCCACCGGCAAUCAUUACUCUUGGC ............((..((....))..))(((.((((((((....))))))))..........(((((...)))))((((((((...((((...))))..)))).)))).........))) ( -32.20) >DroSim_CAF1 36785 120 + 1 CAUUCCCAUACCGAUUUCGAUAGAUUUCGCCUUGUGUGAUCGCUAUCACGCACUCCUCCUCCGGCUGAUUCAGCUGAUUCCGGCCAGGUGACUCACCCACCGGCAAUCAUUACUCUUGGC ............((..((....))..))(((.((((((((....))))))))..........(((((...)))))((((((((...((((...))))..)))).)))).........))) ( -32.20) >DroEre_CAF1 36365 116 + 1 CAUUCCCAUUCCGAUUUCGAUAGAUUUCGCCUUAUGUGAUCGCAAUCACGCACUCCUCCUCCGGCUGAUUCAGCUGAUUCCGGCCAGGUGACUCACCA----CCAAUCAUCACUCUUGGC ............((..((....))..))(((....(((((.((......))...........(((((..((....))...))))).((((......))----))....)))))....))) ( -27.20) >DroYak_CAF1 35558 116 + 1 CAUUUCCAUUCCGAUUUCGAUAGAUUUCGCCUUGUGUGAUCGCUAUCACGCACUCCUCCUCCGGCUGGUUCAGCUUAUUCCGGCCAGGUGACUCACUC----CCAAUCAUCACUCUUAGC ............((((..((..((..((((((((((((((....))))))))..........((((((...........)))))))))))).))..))----..))))............ ( -28.80) >consensus CAUUCCCAUACCGAUUUCGAUAGAUUUCGCCUUGUGUGAUCGCUAUCACGCACUCCUCCUCCGGCUGAUUCAGCUGAUUCCGGCCAGGUGACUCACCC____GCAAUCAUCACUCUUGGC ..........((((....(((.((((..(((.((((((((....)))))))).........((((((...)))))).....)))..((((...)))).......)))))))....)))). (-24.36 = -24.56 + 0.20)

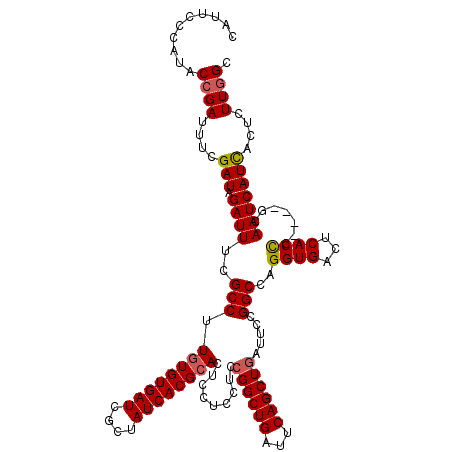
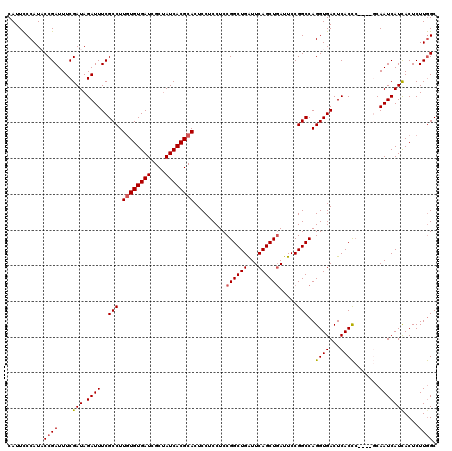
| Location | 4,433,144 – 4,433,260 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.43 |
| Mean single sequence MFE | -18.32 |
| Consensus MFE | -13.44 |
| Energy contribution | -13.45 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.73 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.505189 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4433144 116 + 22224390 CGGCCAGGUGACUCACCC----GCAAUCAUCACUCUUGGCGACCGAUUCCAAUUUCAACUAUUACGAUUCCCAAUAACCGAGGCCAUAUCUCACUUCCAAAUACUUAUUCACACCUUCGC .((((.((((...)))).----.............((((.((.((...................)).)).)))).......))))................................... ( -14.11) >DroSec_CAF1 35513 120 + 1 CGGCCAGGUGACUCACCCACCGGCAAUCAUUACUCUUGGCGACCGAUUCCAAUUCCCACUAUUACGAUUCCCAAUAACCGAGGCCAUAUCUCACUUCCAAAUACUUAUUCACACCUUCGC .((((.((((.......))))((.((((........(((.((...........))))).......)))).)).........))))................................... ( -19.76) >DroSim_CAF1 36865 120 + 1 CGGCCAGGUGACUCACCCACCGGCAAUCAUUACUCUUGGCGACCGAUUCCAAUUCCCACUAUUACGAUUCCCAAUAACCGAGGCCAUAUCUCAUUUCCAAAUACUUAUUCACACCUUCGC .((((.((((.......))))((.((((........(((.((...........))))).......)))).)).........))))................................... ( -19.76) >DroEre_CAF1 36445 100 + 1 CGGCCAGGUGACUCACCA----CCAAUCAUCACUCUUGGCGA----------------CUAUUACGAUUCCCAAUAGCCGAGGCCAUAUCUCACUGCCAAAUACUUAUUCACACUUUCGC .((((.((((......))----))...........(((((..----------------..................)))))))))................................... ( -19.65) >consensus CGGCCAGGUGACUCACCC____GCAAUCAUCACUCUUGGCGACCGAUUCCAAUUCCCACUAUUACGAUUCCCAAUAACCGAGGCCAUAUCUCACUUCCAAAUACUUAUUCACACCUUCGC .((((.((((...))))..................((((.((..........................)))))).......))))................................... (-13.44 = -13.45 + 0.00)

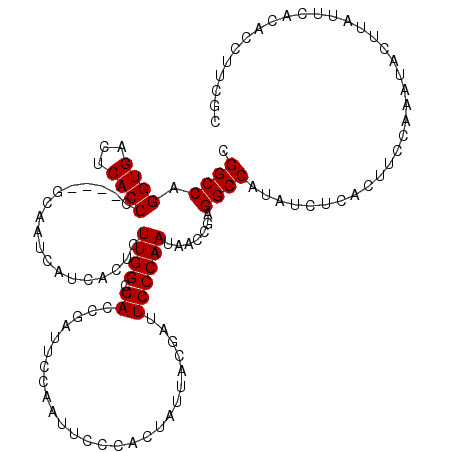
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:11:51 2006