
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,432,026 – 4,432,178 |
| Length | 152 |
| Max. P | 0.958960 |

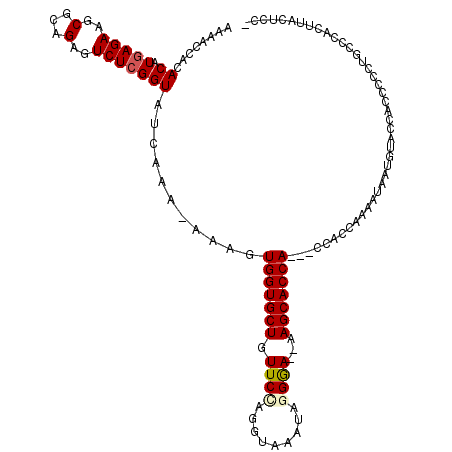
| Location | 4,432,026 – 4,432,140 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.03 |
| Mean single sequence MFE | -25.39 |
| Consensus MFE | -14.68 |
| Energy contribution | -14.96 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.578226 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4432026 114 + 22224390 AAAACCACACAUAAGAAGCACAGAGUCUCGGUAUAAAAAAAAGUGGUGCUGUUCCAGGUAAAUAGGGA--AAGCACCA---CCACCAAAAUAAUGUACCACCCCCUGCCCACUUACUUC- ...........((((..(((..(.((...((((((.......((((((((.((((.........))))--.)))))))---)...........)))))))).)..)))...))))....- ( -24.67) >DroSec_CAF1 34390 101 + 1 AAAACCACACAUGAGAAGCGCAGAGUCUCGGUAUGAAA-AAAGUGGUGCUGUUCGCGGUAAAUAGGGA--AAGCACCA---CCACCAAAAUA------------CUGCCCACUUACUCC- ............((((((.((((.((...(((.(....-)..((((((((.(((.(........).))--))))))))---).)))...)).------------))))...))).))).- ( -25.80) >DroSim_CAF1 35749 101 + 1 AAAACCACACAUGAGAAGCGCAGAGUCUCGGUAUGAAA-AAAGUGGUGCUGUUCUCGGUAAAUAGGGA--AAGCACCA---CCACCAAAACA------------CUGCCCACUUACUCC- ............((((((.((((.((...(((.(....-)..((((((((.(((((........))))--))))))))---).)))...)).------------))))...))).))).- ( -29.20) >DroEre_CAF1 35314 119 + 1 AAAACCACACAUGAGAAGCGCAGAGUCUCGGUAUCAUA-AAAGUGGUGCUGUUCCAGGUAAAUAGGAACAAAGCACCACCACCACCAAAAUAAUGUACCACCCCCUGCCCACCCAUUCCC .................(.((((.(..(.((((.(((.-...(((((((((((((.........)))))..)))))))).............))))))))..).)))).).......... ( -26.03) >DroYak_CAF1 34521 115 + 1 AAAACCCCACAUGAGAAGCGCAGAGUCUCGGUAUCAUA-AAUGUGGUGCUGUUCCAGGUAAAUAUGAAAAAAGCACCA---CCACCAAAAUAAUGUACCACCCCCUGCCCACCCAUUCC- .................(.((((.(..(.((((.(((.-...((((((((.(((...........)))...)))))))---)..........))))))))..).)))).).........- ( -21.24) >consensus AAAACCACACAUGAGAAGCGCAGAGUCUCGGUAUCAAA_AAAGUGGUGCUGUUCCAGGUAAAUAGGGA__AAGCACCA___CCACCAAAAUAAUGUACCACCCCCUGCCCACUUACUCC_ ........((.(((((..(...)..)))))))...........(((((((.((((.........))))...))))))).......................................... (-14.68 = -14.96 + 0.28)

| Location | 4,432,026 – 4,432,140 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.03 |
| Mean single sequence MFE | -38.12 |
| Consensus MFE | -25.40 |
| Energy contribution | -26.20 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.958960 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4432026 114 - 22224390 -GAAGUAAGUGGGCAGGGGGUGGUACAUUAUUUUGGUGG---UGGUGCUU--UCCCUAUUUACCUGGAACAGCACCACUUUUUUUUAUACCGAGACUCUGUGCUUCUUAUGUGUGGUUUU -...(((((.(.((((((..(((((((((.....))))(---((((((((--(((..........)))).)))))))).........)))))...)))))).)..))))).......... ( -36.70) >DroSec_CAF1 34390 101 - 1 -GGAGUAAGUGGGCAG------------UAUUUUGGUGG---UGGUGCUU--UCCCUAUUUACCGCGAACAGCACCACUUU-UUUCAUACCGAGACUCUGCGCUUCUCAUGUGUGGUUUU -.(((.((((..((((------------..(((((((((---((((((((--((.(........).))).)))))))))..-......)))))))..)))))))))))............ ( -33.30) >DroSim_CAF1 35749 101 - 1 -GGAGUAAGUGGGCAG------------UGUUUUGGUGG---UGGUGCUU--UCCCUAUUUACCGAGAACAGCACCACUUU-UUUCAUACCGAGACUCUGCGCUUCUCAUGUGUGGUUUU -.(((.((((..((((------------.((((((((((---((((((((--((.(........).))).)))))))))..-......)))))))).)))))))))))............ ( -36.90) >DroEre_CAF1 35314 119 - 1 GGGAAUGGGUGGGCAGGGGGUGGUACAUUAUUUUGGUGGUGGUGGUGCUUUGUUCCUAUUUACCUGGAACAGCACCACUUU-UAUGAUACCGAGACUCUGCGCUUCUCAUGUGUGGUUUU ....(((((.((((((((..(((((((((((....))))))((((((((..(((((.........)))))))))))))...-.....)))))...)))))).)).))))).......... ( -45.00) >DroYak_CAF1 34521 115 - 1 -GGAAUGGGUGGGCAGGGGGUGGUACAUUAUUUUGGUGG---UGGUGCUUUUUUCAUAUUUACCUGGAACAGCACCACAUU-UAUGAUACCGAGACUCUGCGCUUCUCAUGUGGGGUUUU -...(((((.((((((((..(((((((((.....))))(---(((((((..(((((........))))).))))))))...-.....)))))...)))))).)).))))).......... ( -38.70) >consensus _GGAGUAAGUGGGCAGGGGGUGGUACAUUAUUUUGGUGG___UGGUGCUU__UCCCUAUUUACCUGGAACAGCACCACUUU_UUUCAUACCGAGACUCUGCGCUUCUCAUGUGUGGUUUU ....(((((.(.((((((............((((((((....(((((((...((((........))))..)))))))..........)))))))))))))).)..))))).......... (-25.40 = -26.20 + 0.80)



| Location | 4,432,066 – 4,432,178 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.42 |
| Mean single sequence MFE | -34.88 |
| Consensus MFE | -24.44 |
| Energy contribution | -25.32 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.763057 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4432066 112 + 22224390 AAGUGGUGCUGUUCCAGGUAAAUAGGGA--AAGCACCA---CCACCAAAAUAAUGUACCACCCCCUGCCCACUUACUUC---GGAUGCCUCGAUGGUUCAGCAUUUCCGCGGGCCCUCCU ..((((((((.((((.........))))--.)))))))---)........................((((.........---.((.(((.....))))).((......))))))...... ( -30.60) >DroSec_CAF1 34429 100 + 1 AAGUGGUGCUGUUCGCGGUAAAUAGGGA--AAGCACCA---CCACCAAAAUA------------CUGCCCACUUACUCC---GGAUGGCUCGAUGGUUCGGCAUUUCCGCGGGCCCUCCU ..((((((((.(((.(........).))--))))))))---)..........------------...............---(((.((((((.(((..........))))))))).))). ( -34.30) >DroSim_CAF1 35788 100 + 1 AAGUGGUGCUGUUCUCGGUAAAUAGGGA--AAGCACCA---CCACCAAAACA------------CUGCCCACUUACUCC---GGAUGGCUCGAUGGUUCGGCAUUUCCGCGGGCCCUCCU ..((((((((.(((((........))))--))))))))---)..........------------...............---(((.((((((.(((..........))))))))).))). ( -36.20) >DroEre_CAF1 35353 120 + 1 AAGUGGUGCUGUUCCAGGUAAAUAGGAACAAAGCACCACCACCACCAAAAUAAUGUACCACCCCCUGCCCACCCAUUCCCACGAAUGGCUCGGUGGUUCGGCAUUUCCGCGGGCCCACCU ..(((((((((((((.........)))))..)))))))).................................((((((....))))))...(((((..(.((......)).)..))))). ( -39.60) >DroYak_CAF1 34560 114 + 1 AUGUGGUGCUGUUCCAGGUAAAUAUGAAAAAAGCACCA---CCACCAAAAUAAUGUACCACCCCCUGCCCACCCAUUCC---GGAUGGCUCGGUGGUUCAGCAUUUCCGCGGGCGCUCCU ..((((((((.(((...........)))...)))))))---)..((.......((.((((((....(((...((.....---))..)))..)))))).))((......))))........ ( -33.70) >consensus AAGUGGUGCUGUUCCAGGUAAAUAGGGA__AAGCACCA___CCACCAAAAUAAUGUACCACCCCCUGCCCACUUACUCC___GGAUGGCUCGAUGGUUCGGCAUUUCCGCGGGCCCUCCU ...(((((((.((((.........))))...)))))))............................................(((.((((((.(((..........))))))))).))). (-24.44 = -25.32 + 0.88)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:11:43 2006