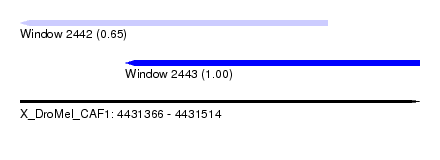
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,431,366 – 4,431,514 |
| Length | 148 |
| Max. P | 0.997314 |

| Location | 4,431,366 – 4,431,480 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.40 |
| Mean single sequence MFE | -26.38 |
| Consensus MFE | -19.94 |
| Energy contribution | -19.70 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.653193 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4431366 114 - 22224390 AACCACCCUGACAUCCAGAUUGCGAUCCGAGCGAAAGACUUGGGCUC-----AAUCGGUUGAUUUCUAUGGCAACUAUCUUUAUCUCCACUGGAAAUAAUAGUACACUUUAUAAG-AGUA .............(((((((((.(.(((((((....).))))))).)-----))))((..(((......(....).......))).))..))))...........((((.....)-))). ( -25.52) >DroSec_CAF1 33734 110 - 1 AACCACCCUGACAUCCAGAUUGCGAUCCGAGCGAAAGACUUGGACUC-----AAUCGGUUGAUUUCUAUGGCAACUAUCUUUAUCUCCUCUGGAAAUAAUAAU----AUUAAUAG-AGUA ...........((.((.(((((.(.(((((((....).))))))).)-----)))))).))..(((((((....)(((..((((.(((...))).))))..))----)...))))-)).. ( -26.40) >DroSim_CAF1 35054 110 - 1 AACCACCCUGACAUCCAGAUUGCGAUCCGAGCGAAAGACUUGGACUC-----AAUCGGUUGAUUUCUAUGGCAACUAUCUUUAUCUCCACUGGAAAUAAUAAU----UUUAAUAG-AGUA ....((.(((.((.((.(((((.(.(((((((....).))))))).)-----)))))).))((((((((((...............))).)))))))......----.....)))-.)). ( -25.96) >DroEre_CAF1 34716 107 - 1 AACCACCCUGACAUCCAGAUUGCGAUCCGAGCGAAAGACUUGGGCUCAAGCCAAUCGGUUGAUUUCUAUGGCAACUAUCUUUAUCUGCACUGGAAAUAAU-------------AUAAACU .............(((((..((((((((((((....).)))))).))..((((...((......))..))))..............))))))))......-------------....... ( -25.50) >DroYak_CAF1 33911 119 - 1 AACCACCCUGGCAUCCAGAUUGCGAUCCGAGCGAAAGACUUGGGCUCAAGUCAAUCGGUUGAUUUCUAUGGAAACUAUCUUUAUCUGCACUGGAAAUAAUAGUGCAC-UUAAUAAAAAUU ..(((..((((...)))).....(((((((((....).)))))).))(((((((....)))))))...))).........(((..(((((((.......))))))).-.)))........ ( -28.50) >consensus AACCACCCUGACAUCCAGAUUGCGAUCCGAGCGAAAGACUUGGGCUC_____AAUCGGUUGAUUUCUAUGGCAACUAUCUUUAUCUCCACUGGAAAUAAUAAU_____UUAAUAG_AGUA .............(((((.....(((((((((....).)))))).))..........(..(((......(....).......)))..).))))).......................... (-19.94 = -19.70 + -0.24)


| Location | 4,431,405 – 4,431,514 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.20 |
| Mean single sequence MFE | -35.46 |
| Consensus MFE | -29.45 |
| Energy contribution | -29.80 |
| Covariance contribution | 0.35 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.84 |
| SVM RNA-class probability | 0.997314 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4431405 109 - 22224390 CCUAACGGUUGCCUGAA------UUCAAACAGGAGUUCCGAACCACCCUGACAUCCAGAUUGCGAUCCGAGCGAAAGACUUGGGCUC-----AAUCGGUUGAUUUCUAUGGCAACUAUCU ......(((((((.(((------.((((.((((.((........))))))....((.(((((.(.(((((((....).))))))).)-----)))))))))).)))...))))))).... ( -38.50) >DroSec_CAF1 33769 115 - 1 CCUAACGGUUGCCUGAAUUCGAAUUCAAACAGGAGUUCCGAACCACCCUGACAUCCAGAUUGCGAUCCGAGCGAAAGACUUGGACUC-----AAUCGGUUGAUUUCUAUGGCAACUAUCU ......(((((((.(((.((((((((......))))))..((((....((.....))(((((.(.(((((((....).))))))).)-----)))))))))).)))...))))))).... ( -38.90) >DroSim_CAF1 35089 115 - 1 CCUAACGGUUGCCUGAAUUCGAAUUCAAACAGGAGUUCCGAACCACCCUGACAUCCAGAUUGCGAUCCGAGCGAAAGACUUGGACUC-----AAUCGGUUGAUUUCUAUGGCAACUAUCU ......(((((((.(((.((((((((......))))))..((((....((.....))(((((.(.(((((((....).))))))).)-----)))))))))).)))...))))))).... ( -38.90) >DroEre_CAF1 34743 114 - 1 CCUAACGGUUGCCUUAA------UUCAAACAGGAGUUCCGAACCACCCUGACAUCCAGAUUGCGAUCCGAGCGAAAGACUUGGGCUCAAGCCAAUCGGUUGAUUUCUAUGGCAACUAUCU ......(((((((....------......((((.((........)))))).((.((.(((((((((((((((....).)))))).))..).))))))).))........))))))).... ( -33.10) >DroYak_CAF1 33950 114 - 1 CCUAACGGUUGCCUUAA------UUCAAACAGGAGUUUCGAACCACCCUGGCAUCCAGAUUGCGAUCCGAGCGAAAGACUUGGGCUCAAGUCAAUCGGUUGAUUUCUAUGGAAACUAUCU .....((((((.(((..------......((((.((........))))))(((.......)))(((((((((....).)))))).))))).))))))............(....)..... ( -27.90) >consensus CCUAACGGUUGCCUGAA______UUCAAACAGGAGUUCCGAACCACCCUGACAUCCAGAUUGCGAUCCGAGCGAAAGACUUGGGCUC_____AAUCGGUUGAUUUCUAUGGCAACUAUCU ......((((((((((((....)))))...(((((((((((......(((.....))).....(((((((((....).)))))).)).......))))..)))))))..))))))).... (-29.45 = -29.80 + 0.35)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:11:41 2006