
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,430,007 – 4,430,127 |
| Length | 120 |
| Max. P | 0.707105 |

| Location | 4,430,007 – 4,430,127 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.83 |
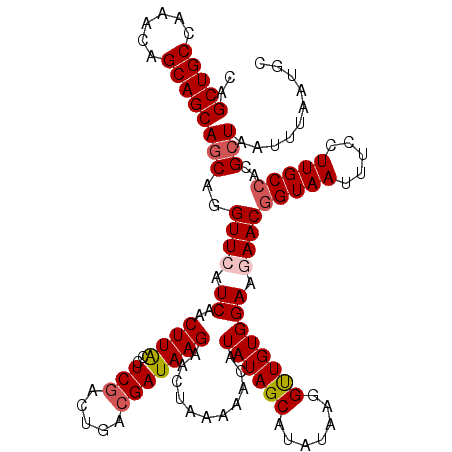
| Mean single sequence MFE | -28.06 |
| Consensus MFE | -23.72 |
| Energy contribution | -24.24 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.707105 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4430007 120 + 22224390 CAGCUGCCAAACAGCAGCAACAGGUUCAUCAACUUACCUCGACUGACGAUAAGAGCUAAAAACAUAUAGCAUAUAAGGUUGUGGAAGAACGGUAAUUUUCUUGCCACGCUAAUUUAAUGC ..(((((......))))).....((((.((.((..((((((.....))).....((((........))))......))).)).)).))))(((((.....)))))............... ( -26.70) >DroSec_CAF1 32371 120 + 1 CAGCUGCCAAACAGCAGCAGCAGGUUCAUCAACUUGCCUCGACUGACGAUAAGAAUUUAAAACAUAUAGCAUAUAAGGCUGUGGAAGAACGGUAAUUUCCUUGCCACGCUAAUUUAAUGC ..(((((......)))))(((..((((.((..((((..(((.....)))))))...........((((((.......)))))))).))))(((((.....)))))..))).......... ( -29.00) >DroSim_CAF1 33697 120 + 1 CAGCUGCCAAACAGCAGCAGCAGGUUCAUCAACUUGCCUCGACUGACGAUAAGAAUUUAAAACAUAUAGCAUAUAAGGCUGUGGAAGAACGGUAAUUUCCUUGCCACGCUAAUUUAAUGC ..(((((......)))))(((..((((.((..((((..(((.....)))))))...........((((((.......)))))))).))))(((((.....)))))..))).......... ( -29.00) >DroEre_CAF1 33419 120 + 1 CAGCUGCCAAACGGCAGCAGCAGGUUCAUCAACUUACCUCGCCUGACGAUAAGAACUAAAAACAUAUAGCAUAUAAGGUUGUGGAAAAACGGUAAUUUCCUUGCCACGCUAAUUUAAUGC ..((((((....))))))(((...((((.((((((...(((.....)))......(((........)))......)))))))))).....(((((.....)))))..))).......... ( -27.00) >DroYak_CAF1 32757 120 + 1 CAGCUGCCAAACGGCAGCAGCAGGUUCAUCAACUUACCUCCACUGACGAUAAGAGCUAAAAACAUAUAGCAUAUAAGGUUGUGGAAAAACUGUAAUUUCCUUGCCACGCUAAUUUAAUGC ..((((((....))))))(((((((..........))))...............((((........))))......(((...(((((........)))))..)))..))).......... ( -28.60) >consensus CAGCUGCCAAACAGCAGCAGCAGGUUCAUCAACUUACCUCGACUGACGAUAAGAACUAAAAACAUAUAGCAUAUAAGGUUGUGGAAGAACGGUAAUUUCCUUGCCACGCUAAUUUAAUGC ..(((((......)))))(((..((((.((..((((..(((.....)))))))...........((((((.......)))))))).))))(((((.....)))))..))).......... (-23.72 = -24.24 + 0.52)

| Location | 4,430,007 – 4,430,127 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.83 |
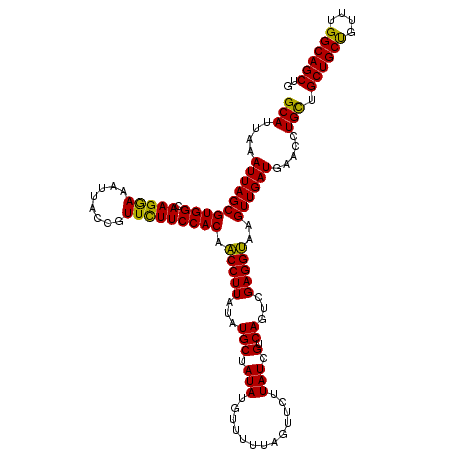
| Mean single sequence MFE | -31.57 |
| Consensus MFE | -31.60 |
| Energy contribution | -30.32 |
| Covariance contribution | -1.28 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.03 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.634522 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4430007 120 - 22224390 GCAUUAAAUUAGCGUGGCAAGAAAAUUACCGUUCUUCCACAACCUUAUAUGCUAUAUGUUUUUAGCUCUUAUCGUCAGUCGAGGUAAGUUGAUGAACCUGUUGCUGCUGUUUGGCAGCUG (((....((((((((((.(((((........))))))))).(((......((((........)))).....(((.....))))))..)))))).....))).((((((....)))))).. ( -31.00) >DroSec_CAF1 32371 120 - 1 GCAUUAAAUUAGCGUGGCAAGGAAAUUACCGUUCUUCCACAGCCUUAUAUGCUAUAUGUUUUAAAUUCUUAUCGUCAGUCGAGGCAAGUUGAUGAACCUGCUGCUGCUGUUUGGCAGCUG (((....((((((((((.(((((........))))))))).(((((...(((.(((.............))).).))...)))))..)))))).....))).((((((....)))))).. ( -32.42) >DroSim_CAF1 33697 120 - 1 GCAUUAAAUUAGCGUGGCAAGGAAAUUACCGUUCUUCCACAGCCUUAUAUGCUAUAUGUUUUAAAUUCUUAUCGUCAGUCGAGGCAAGUUGAUGAACCUGCUGCUGCUGUUUGGCAGCUG (((....((((((((((.(((((........))))))))).(((((...(((.(((.............))).).))...)))))..)))))).....))).((((((....)))))).. ( -32.42) >DroEre_CAF1 33419 120 - 1 GCAUUAAAUUAGCGUGGCAAGGAAAUUACCGUUUUUCCACAACCUUAUAUGCUAUAUGUUUUUAGUUCUUAUCGUCAGGCGAGGUAAGUUGAUGAACCUGCUGCUGCCGUUUGGCAGCUG ((.........((.((((..(((((........)))))............((((........)))).......)))).)).((((..........)))))).((((((....)))))).. ( -30.80) >DroYak_CAF1 32757 120 - 1 GCAUUAAAUUAGCGUGGCAAGGAAAUUACAGUUUUUCCACAACCUUAUAUGCUAUAUGUUUUUAGCUCUUAUCGUCAGUGGAGGUAAGUUGAUGAACCUGCUGCUGCCGUUUGGCAGCUG (((....((((((((((.(((((........))))))))).(((((...(((.(((.((.....))...))).).))...)))))..)))))).....))).((((((....)))))).. ( -31.20) >consensus GCAUUAAAUUAGCGUGGCAAGGAAAUUACCGUUCUUCCACAACCUUAUAUGCUAUAUGUUUUUAGUUCUUAUCGUCAGUCGAGGUAAGUUGAUGAACCUGCUGCUGCUGUUUGGCAGCUG (((....((((((((((.(((((........))))))))).(((((...(((.(((.............))).).))...)))))..)))))).....))).((((((....)))))).. (-31.60 = -30.32 + -1.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:11:36 2006