
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,427,479 – 4,427,639 |
| Length | 160 |
| Max. P | 0.998760 |

| Location | 4,427,479 – 4,427,599 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.33 |
| Mean single sequence MFE | -41.60 |
| Consensus MFE | -39.02 |
| Energy contribution | -38.78 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.94 |
| SVM decision value | 3.21 |
| SVM RNA-class probability | 0.998760 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4427479 120 - 22224390 UCCUUUUGUCUUUGUCGAUGGCCAACUGCAUCGACGACGGCUGCUGGCGGUGCUGCUGCGCCAAUCUUCAAUGGAAGCCAAAGCUAAAGCUAAAGCCAAUACAGCGCCAAAUGGGCAGGC .(((.((((..(((((((((........))))))))).((((..(((((((((....)))))..(((.....))).)))).(((....)))..))))...)))).(((.....)))))). ( -41.90) >DroSec_CAF1 29884 120 - 1 UCCUUUUGUCUUUGUCGAUGGCCAACUGCAUCGACGACGGCUGCUGGCGGUGCUGCUGCGCCAAUCUUCAAUGGAAGCCAAAGCUAAAGCUAAAGCCAAUACAACGCCAAAUGGGCAGGC .(((.((((..(((((((((........))))))))).((((..(((((((((....)))))..(((.....))).)))).(((....)))..))))...)))).(((.....)))))). ( -42.20) >DroSim_CAF1 31192 120 - 1 UCCUUUUGUCUUUGUCGAUGGCCAACUGCAUCGACGACGGCUGCUGGCGGUGCUGCUGCGCCAAUCUUCAAUGGAAGCCAAAGCCAAAGCUAAAGCCAAUACAACGCCAAAUGGGCAGGC .(((.(((.(((((((((((........)))))))...((((..(((((((((....)))))..(((.....))).)))).))))......)))).)))......(((.....)))))). ( -42.50) >DroEre_CAF1 30857 120 - 1 UCCUUUUGUCUUUGUCGAUGGCCAACUGCAUCGACAACGGCUGCUGGCGGUGCUGCUGCGCCAAUCUUCAAUGGAAGCCAACGCUAAAGCUAAAGCCAAUACAACGCCAAAUGGGCAGGC .(((.((((..(((((((((........))))))))).(((((.(((((((((....)))))..(((.....))).)))).)((....))...))))...)))).(((.....)))))). ( -41.60) >DroYak_CAF1 30404 114 - 1 UCCUUUUGUCUUUGUCGAUGGCCAACUGCAUCGACAACGGCUGCUGGCGGUGCUGCUGCGCCAAUCUUCAAUGGAAGCCA------AAGCUAAAGCCAAUACAACGCCAAAUGGGACUUC ((((((((.(.(((((((((........))))))))).((((..(((((((((....)))))...((((....))))...------..)))).))))........).)))).)))).... ( -39.80) >consensus UCCUUUUGUCUUUGUCGAUGGCCAACUGCAUCGACGACGGCUGCUGGCGGUGCUGCUGCGCCAAUCUUCAAUGGAAGCCAAAGCUAAAGCUAAAGCCAAUACAACGCCAAAUGGGCAGGC (((.((((.(.(((((((((........))))))))).((((..(((((((((....)))))...((((....))))...........)))).))))........).)))).)))..... (-39.02 = -38.78 + -0.24)

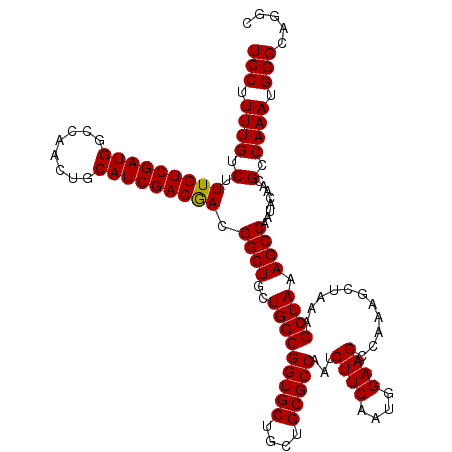

| Location | 4,427,519 – 4,427,639 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -33.50 |
| Consensus MFE | -31.90 |
| Energy contribution | -31.66 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.967200 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4427519 120 + 22224390 UGGCUUCCAUUGAAGAUUGGCGCAGCAGCACCGCCAGCAGCCGUCGUCGAUGCAGUUGGCCAUCGACAAAGACAAAAGGAAAACACUCCAUGAAUCUUUAAUGAAGAAUCACCAGUUACC (((((((.(((((((((((((((.((......))..)).)))(((((((((((.....).)))))))...)))....(((......)))...)))))))))))))).....)))...... ( -34.60) >DroSec_CAF1 29924 119 + 1 UGGCUUCCAUUGAAGAUUGGCGCAGCAGCACCGCCAGCAGCCGUCGUCGAUGCAGUUGGCCAUCGACAAAGACAAAAGGAAAACACACCAUGAAUCUUUAAUGAAGAAUCACCAGUUAC- (((((((.(((((((((((((((.((......))..)).)))(((((((((((.....).)))))))...)))....((........))...)))))))))))))).....))).....- ( -32.60) >DroSim_CAF1 31232 120 + 1 UGGCUUCCAUUGAAGAUUGGCGCAGCAGCACCGCCAGCAGCCGUCGUCGAUGCAGUUGGCCAUCGACAAAGACAAAAGGAAAACACACCAUGAAUCUUUAAUGAAGAAUCACCAGUUACC (((((((.(((((((((((((((.((......))..)).)))(((((((((((.....).)))))))...)))....((........))...)))))))))))))).....)))...... ( -32.60) >DroEre_CAF1 30897 120 + 1 UGGCUUCCAUUGAAGAUUGGCGCAGCAGCACCGCCAGCAGCCGUUGUCGAUGCAGUUGGCCAUCGACAAAGACAAAAGGAAAACACUCCAUGAAUCUUUAAUGAAGAAUCACCAGUUACC (((((((.(((((((((((((.((((.(((.((.((((....)))).)).))).)))))))................(((......)))...)))))))))))))).....)))...... ( -35.30) >DroYak_CAF1 30438 120 + 1 UGGCUUCCAUUGAAGAUUGGCGCAGCAGCACCGCCAGCAGCCGUUGUCGAUGCAGUUGGCCAUCGACAAAGACAAAAGGAAAACACUACAUGAAUCUUUAAUGAAGAAUCACCAGUUACC (((((((.(((((((((((((((.((......))..)).))).((((((((((.....).))))))))).......................)))))))))))))).....)))...... ( -32.40) >consensus UGGCUUCCAUUGAAGAUUGGCGCAGCAGCACCGCCAGCAGCCGUCGUCGAUGCAGUUGGCCAUCGACAAAGACAAAAGGAAAACACUCCAUGAAUCUUUAAUGAAGAAUCACCAGUUACC (((((((.(((((((((((((((.((......))..)).)))(((((((((((.....).)))))))...)))...................)))))))))))))).....)))...... (-31.90 = -31.66 + -0.24)

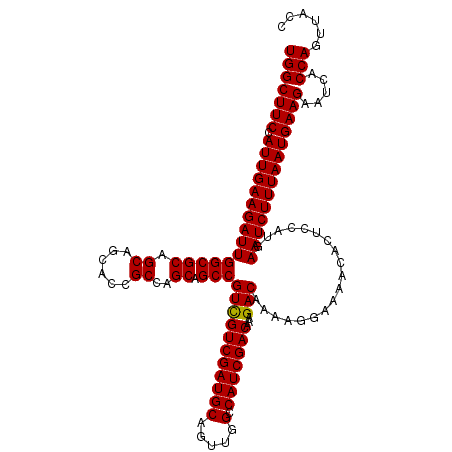

| Location | 4,427,519 – 4,427,639 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -38.80 |
| Consensus MFE | -37.36 |
| Energy contribution | -37.12 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.946213 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4427519 120 - 22224390 GGUAACUGGUGAUUCUUCAUUAAAGAUUCAUGGAGUGUUUUCCUUUUGUCUUUGUCGAUGGCCAACUGCAUCGACGACGGCUGCUGGCGGUGCUGCUGCGCCAAUCUUCAAUGGAAGCCA ((((.(((((((....)))).((((((....((((....))))....))))))(((((((........)))))))..))).))))((((((((....)))))..(((.....))).))). ( -39.80) >DroSec_CAF1 29924 119 - 1 -GUAACUGGUGAUUCUUCAUUAAAGAUUCAUGGUGUGUUUUCCUUUUGUCUUUGUCGAUGGCCAACUGCAUCGACGACGGCUGCUGGCGGUGCUGCUGCGCCAAUCUUCAAUGGAAGCCA -......(((.....((((((.((((((.............((....(((.(((((((((........))))))))).)))....)).(((((....))))))))))).)))))).))). ( -37.00) >DroSim_CAF1 31232 120 - 1 GGUAACUGGUGAUUCUUCAUUAAAGAUUCAUGGUGUGUUUUCCUUUUGUCUUUGUCGAUGGCCAACUGCAUCGACGACGGCUGCUGGCGGUGCUGCUGCGCCAAUCUUCAAUGGAAGCCA (((((((.((((.((((.....)))).)))).).))...........(((.(((((((((........))))))))).)))))))((((((((....)))))..(((.....))).))). ( -37.90) >DroEre_CAF1 30897 120 - 1 GGUAACUGGUGAUUCUUCAUUAAAGAUUCAUGGAGUGUUUUCCUUUUGUCUUUGUCGAUGGCCAACUGCAUCGACAACGGCUGCUGGCGGUGCUGCUGCGCCAAUCUUCAAUGGAAGCCA ((((....((((.((((.....)))).))))((((....))))....(((.(((((((((........))))))))).)))))))((((((((....)))))..(((.....))).))). ( -40.00) >DroYak_CAF1 30438 120 - 1 GGUAACUGGUGAUUCUUCAUUAAAGAUUCAUGUAGUGUUUUCCUUUUGUCUUUGUCGAUGGCCAACUGCAUCGACAACGGCUGCUGGCGGUGCUGCUGCGCCAAUCUUCAAUGGAAGCCA ((((((((((((.((((.....)))).)))).))))...........(((.(((((((((........))))))))).)))))))((((((((....)))))..(((.....))).))). ( -39.30) >consensus GGUAACUGGUGAUUCUUCAUUAAAGAUUCAUGGAGUGUUUUCCUUUUGUCUUUGUCGAUGGCCAACUGCAUCGACGACGGCUGCUGGCGGUGCUGCUGCGCCAAUCUUCAAUGGAAGCCA .......(((.....((((((.((((((.............((....(((.(((((((((........))))))))).)))....)).(((((....))))))))))).)))))).))). (-37.36 = -37.12 + -0.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:11:22 2006