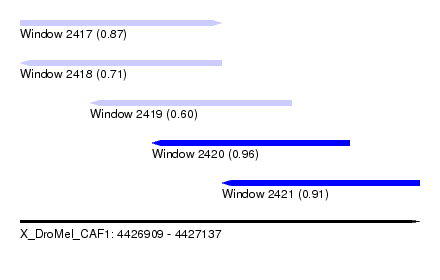
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,426,909 – 4,427,137 |
| Length | 228 |
| Max. P | 0.964453 |

| Location | 4,426,909 – 4,427,024 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 92.17 |
| Mean single sequence MFE | -37.48 |
| Consensus MFE | -31.70 |
| Energy contribution | -32.57 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.865083 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4426909 115 + 22224390 CCCUCGACGCGUCUUGCCUCUUUCUUGACUGGCACGAAGAUUGGAUUGGAUUGGGAUUUCAAGAAGAAAUCAAGCCAGCCAACUGUCAGUGCCGAGGCAUGGCAAAGCAUUUUAA ........(((((.((((((((....))..(((((..((.((((.((((.....((((((.....))))))...))))))))))....))))))))))).)))...))....... ( -37.10) >DroSec_CAF1 29294 115 + 1 CCCGCGCCGCGUCUUGCCUCUUAGUUGACUGGCACGAAGAUUGGAUUGGAUUGGGAUUUCAAGAAGAAAUCAAGCCAGCCAACUGUCAGUGCCGAGGCAUGGCAAAGCAUUUUAA .....((...(((.((((((..((....))(((((..((.((((.((((.....((((((.....))))))...))))))))))....))))))))))).)))...))....... ( -40.50) >DroSim_CAF1 30600 115 + 1 CCCGCGCCGCGUCUUGCCUCUUAGUUGACUGGCACGAAGAUUGGAUUGGAUUGGGAUUUCAAGAAGAAAUCAAGCCAGCCAACUGUCAGUGCCGAGGCAUGGCAAAGCAUUUUAA .....((...(((.((((((..((....))(((((..((.((((.((((.....((((((.....))))))...))))))))))....))))))))))).)))...))....... ( -40.50) >DroYak_CAF1 29843 105 + 1 CCCUCG----------UUUCUGGCUUGACUGGCACGAAGAUUGGAUUGGAUUGGGAUUUCAAGAAGAAAUCAAGCCAGCCAACUGUCAGUGCCGAGGCAUGGCAAAGCAUUUUAA .....(----------(((.((.(.((.(((((((..((.((((.((((.....((((((.....))))))...))))))))))....))))).)).)).).))))))....... ( -31.80) >consensus CCCGCGCCGCGUCUUGCCUCUUACUUGACUGGCACGAAGAUUGGAUUGGAUUGGGAUUUCAAGAAGAAAUCAAGCCAGCCAACUGUCAGUGCCGAGGCAUGGCAAAGCAUUUUAA ..........(((.((((((..........(((((..((.((((.((((.....((((((.....))))))...))))))))))....))))))))))).)))............ (-31.70 = -32.57 + 0.87)



| Location | 4,426,909 – 4,427,024 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 92.17 |
| Mean single sequence MFE | -34.85 |
| Consensus MFE | -27.95 |
| Energy contribution | -30.20 |
| Covariance contribution | 2.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.709755 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4426909 115 - 22224390 UUAAAAUGCUUUGCCAUGCCUCGGCACUGACAGUUGGCUGGCUUGAUUUCUUCUUGAAAUCCCAAUCCAAUCCAAUCUUCGUGCCAGUCAAGAAAGAGGCAAGACGCGUCGAGGG ..................(((((((((.((..(((((.(((...((((((.....)))))).....)))..)))))..)))))))..........((.((.....)).)))))). ( -34.20) >DroSec_CAF1 29294 115 - 1 UUAAAAUGCUUUGCCAUGCCUCGGCACUGACAGUUGGCUGGCUUGAUUUCUUCUUGAAAUCCCAAUCCAAUCCAAUCUUCGUGCCAGUCAACUAAGAGGCAAGACGCGGCGCGGG ......(((.((((..(((((((((((.((..(((((.(((...((((((.....)))))).....)))..)))))..)))))))((....))..))))))....)))).))).. ( -38.30) >DroSim_CAF1 30600 115 - 1 UUAAAAUGCUUUGCCAUGCCUCGGCACUGACAGUUGGCUGGCUUGAUUUCUUCUUGAAAUCCCAAUCCAAUCCAAUCUUCGUGCCAGUCAACUAAGAGGCAAGACGCGGCGCGGG ......(((.((((..(((((((((((.((..(((((.(((...((((((.....)))))).....)))..)))))..)))))))((....))..))))))....)))).))).. ( -38.30) >DroYak_CAF1 29843 105 - 1 UUAAAAUGCUUUGCCAUGCCUCGGCACUGACAGUUGGCUGGCUUGAUUUCUUCUUGAAAUCCCAAUCCAAUCCAAUCUUCGUGCCAGUCAAGCCAGAAA----------CGAGGG ..................(((((((((.((..(((((.(((...((((((.....)))))).....)))..)))))..)))))))..((......))..----------.)))). ( -28.60) >consensus UUAAAAUGCUUUGCCAUGCCUCGGCACUGACAGUUGGCUGGCUUGAUUUCUUCUUGAAAUCCCAAUCCAAUCCAAUCUUCGUGCCAGUCAACUAAGAGGCAAGACGCGGCGAGGG ........(((((((.(((((((((((.((..(((((.(((...((((((.....)))))).....)))..)))))..)))))))..........))))))......))))))). (-27.95 = -30.20 + 2.25)


| Location | 4,426,949 – 4,427,064 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 99.57 |
| Mean single sequence MFE | -28.52 |
| Consensus MFE | -28.40 |
| Energy contribution | -28.40 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.83 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.602893 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4426949 115 - 22224390 GCCGUUGGCCUAAAAUAAUGGCCGCAAGAAAAUUUCUAUAUUAAAAUGCUUUGCCAUGCCUCGGCACUGACAGUUGGCUGGCUUGAUUUCUUCUUGAAAUCCCAAUCCAAUCCAA ((((..(((........(((((.(((....(((......)))....)))...)))))))).)))).......(((((.(((...((((((.....)))))))))..))))).... ( -28.40) >DroSec_CAF1 29334 115 - 1 GCCGUUGGCCUAAAAUAAUGGCCGCAAGUAAAUUUCUAUAUUAAAAUGCUUUGCCAUGCCUCGGCACUGACAGUUGGCUGGCUUGAUUUCUUCUUGAAAUCCCAAUCCAAUCCAA ((((..(((........(((((.(((..(.(((......))).)..)))...)))))))).)))).......(((((.(((...((((((.....)))))))))..))))).... ( -28.90) >DroSim_CAF1 30640 115 - 1 GCCGUUGGCCUAAAAUAAUGGCCGCAAGAAAAUUUCUAUAUUAAAAUGCUUUGCCAUGCCUCGGCACUGACAGUUGGCUGGCUUGAUUUCUUCUUGAAAUCCCAAUCCAAUCCAA ((((..(((........(((((.(((....(((......)))....)))...)))))))).)))).......(((((.(((...((((((.....)))))))))..))))).... ( -28.40) >DroYak_CAF1 29873 115 - 1 GCCGUUGGCCUAAAAUAAUGGCCGCAAGAAAAUUUCUAUAUUAAAAUGCUUUGCCAUGCCUCGGCACUGACAGUUGGCUGGCUUGAUUUCUUCUUGAAAUCCCAAUCCAAUCCAA ((((..(((........(((((.(((....(((......)))....)))...)))))))).)))).......(((((.(((...((((((.....)))))))))..))))).... ( -28.40) >consensus GCCGUUGGCCUAAAAUAAUGGCCGCAAGAAAAUUUCUAUAUUAAAAUGCUUUGCCAUGCCUCGGCACUGACAGUUGGCUGGCUUGAUUUCUUCUUGAAAUCCCAAUCCAAUCCAA ((((..(((........(((((.(((....(((......)))....)))...)))))))).)))).......(((((.(((...((((((.....)))))))))..))))).... (-28.40 = -28.40 + 0.00)

| Location | 4,426,984 – 4,427,097 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 99.65 |
| Mean single sequence MFE | -38.30 |
| Consensus MFE | -38.30 |
| Energy contribution | -38.30 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.69 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.964453 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4426984 113 - 22224390 GCUGACAUGGGCCUCGACUUCGGGCCGUUGUAGGCCGUUGGCCUAAAAUAAUGGCCGCAAGAAAAUUUCUAUAUUAAAAUGCUUUGCCAUGCCUCGGCACUGACAGUUGGCUG ((..((.((((((.((....))))))((((.((((....((((.........))))(((((...((((........))))..)))))...)))))))).....))))..)).. ( -38.30) >DroSec_CAF1 29369 113 - 1 GCUGACAUGGGCCUCGACUUCGGGCCGUUGUAGGCCGUUGGCCUAAAAUAAUGGCCGCAAGUAAAUUUCUAUAUUAAAAUGCUUUGCCAUGCCUCGGCACUGACAGUUGGCUG ((..((.((((((.((....))))))((((.((((....((((.........))))(((((...((((........))))..)))))...)))))))).....))))..)).. ( -38.30) >DroSim_CAF1 30675 113 - 1 GCUGACAUGGGCCUCGACUUCGGGCCGUUGUAGGCCGUUGGCCUAAAAUAAUGGCCGCAAGAAAAUUUCUAUAUUAAAAUGCUUUGCCAUGCCUCGGCACUGACAGUUGGCUG ((..((.((((((.((....))))))((((.((((....((((.........))))(((((...((((........))))..)))))...)))))))).....))))..)).. ( -38.30) >DroEre_CAF1 30305 113 - 1 GCUGACAUGGGCCUCGACUUCGGGCCGUUGUAGGCCGUUGGCCUAAAAUAAUGGCCGCAAGAAAAUUUCUAUAUUAAAAUGCUUUGCCAUGCCUCGGCACUGACAGUUGGCUG ((..((.((((((.((....))))))((((.((((....((((.........))))(((((...((((........))))..)))))...)))))))).....))))..)).. ( -38.30) >DroYak_CAF1 29908 113 - 1 GCUGACAUGGGCCUCGACUUCGGGCCGUUGUAGGCCGUUGGCCUAAAAUAAUGGCCGCAAGAAAAUUUCUAUAUUAAAAUGCUUUGCCAUGCCUCGGCACUGACAGUUGGCUG ((..((.((((((.((....))))))((((.((((....((((.........))))(((((...((((........))))..)))))...)))))))).....))))..)).. ( -38.30) >consensus GCUGACAUGGGCCUCGACUUCGGGCCGUUGUAGGCCGUUGGCCUAAAAUAAUGGCCGCAAGAAAAUUUCUAUAUUAAAAUGCUUUGCCAUGCCUCGGCACUGACAGUUGGCUG ((..((.((((((.((....))))))((((.((((....((((.........))))(((((...((((........))))..)))))...)))))))).....))))..)).. (-38.30 = -38.30 + 0.00)

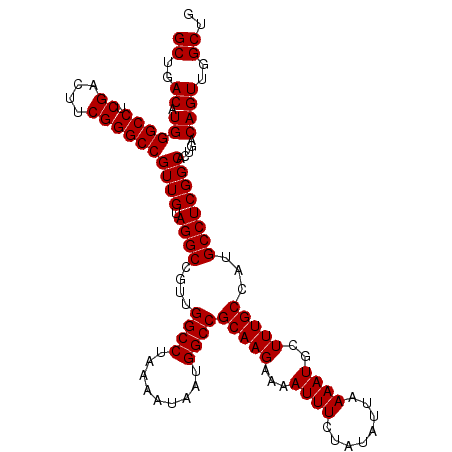
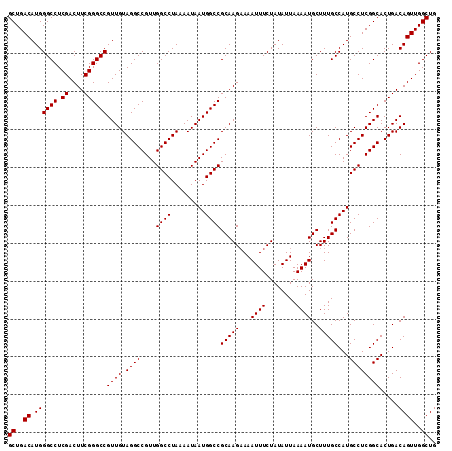
| Location | 4,427,024 – 4,427,137 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 92.53 |
| Mean single sequence MFE | -30.51 |
| Consensus MFE | -27.06 |
| Energy contribution | -26.78 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.913026 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4427024 113 - 22224390 CUGGUAAAACACCAUACAUAUUUUUUUUUGCCUGUAUGCUGCUGACAUGGGCCUCGACUUCGGGCCGUUGUAGGCCGUUGGCCUAAAAUAAUGGCCGCAAGAAAAUUUCUAUA .((((.....))))........(((((((((......((.(((.(((..((((.((....))))))..))).))).)).((((.........)))))))))))))........ ( -34.30) >DroSec_CAF1 29409 113 - 1 CUGUUAAAACCCCAUAUAUCUUUUUUUUUGCCUGUAGGCUGCUGACAUGGGCCUCGACUUCGGGCCGUUGUAGGCCGUUGGCCUAAAAUAAUGGCCGCAAGUAAAUUUCUAUA ..........................(((((.(((.((((....(((..((((.((....))))))..))).))))...((((.........))))))).)))))........ ( -31.50) >DroSim_CAF1 30715 112 - 1 CUGUUAAAACCCCAUAUAUU-UUUUUUCUGCCUGUAUGCUGCUGACAUGGGCCUCGACUUCGGGCCGUUGUAGGCCGUUGGCCUAAAAUAAUGGCCGCAAGAAAAUUUCUAUA ..............((((..-..((((((((......((.(((.(((..((((.((....))))))..))).))).)).((((.........)))))).))))))....)))) ( -29.90) >DroEre_CAF1 30345 109 - 1 CUGUUAAUACUCUAUAUAUU-U--UU-UUCCCUGUAUGCUGCUGACAUGGGCCUCGACUUCGGGCCGUUGUAGGCCGUUGGCCUAAAAUAAUGGCCGCAAGAAAAUUUCUAUA ....................-.--((-(((..(((..((.(((.(((..((((.((....))))))..))).))).)).((((.........))))))).)))))........ ( -27.40) >DroYak_CAF1 29948 110 - 1 CUGGUAAUAAUCUAUACAUU-U--UUUUUACCUGUAUGCUGCUGACAUGGGCCUCGACUUCGGGCCGUUGUAGGCCGUUGGCCUAAAAUAAUGGCCGCAAGAAAAUUUCUAUA ..(((((.............-.--...)))))....(((.....(((..((((.((....))))))..))).((((((((........))))))))))).............. ( -29.43) >consensus CUGUUAAAACCCCAUAUAUU_UUUUUUUUGCCUGUAUGCUGCUGACAUGGGCCUCGACUUCGGGCCGUUGUAGGCCGUUGGCCUAAAAUAAUGGCCGCAAGAAAAUUUCUAUA ............((((((..............)))))).(((..(((..((((.((....))))))..))).((((((((........))))))))))).............. (-27.06 = -26.78 + -0.28)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:11:19 2006